Snorkeling
This repository stores data and code to scale up the extraction of biomedical relationships (i.e. Disease-Gene associations, Compounds binding to Genes, Gene-Gene interactions etc.) from the Pubmed Abstracts.
Depreciation Note
An updated version of this project can be found at: greenelab/snorkeling-full-text. New changes pertaining to the repository can be found at the link provided previously.
Quick Synopsis
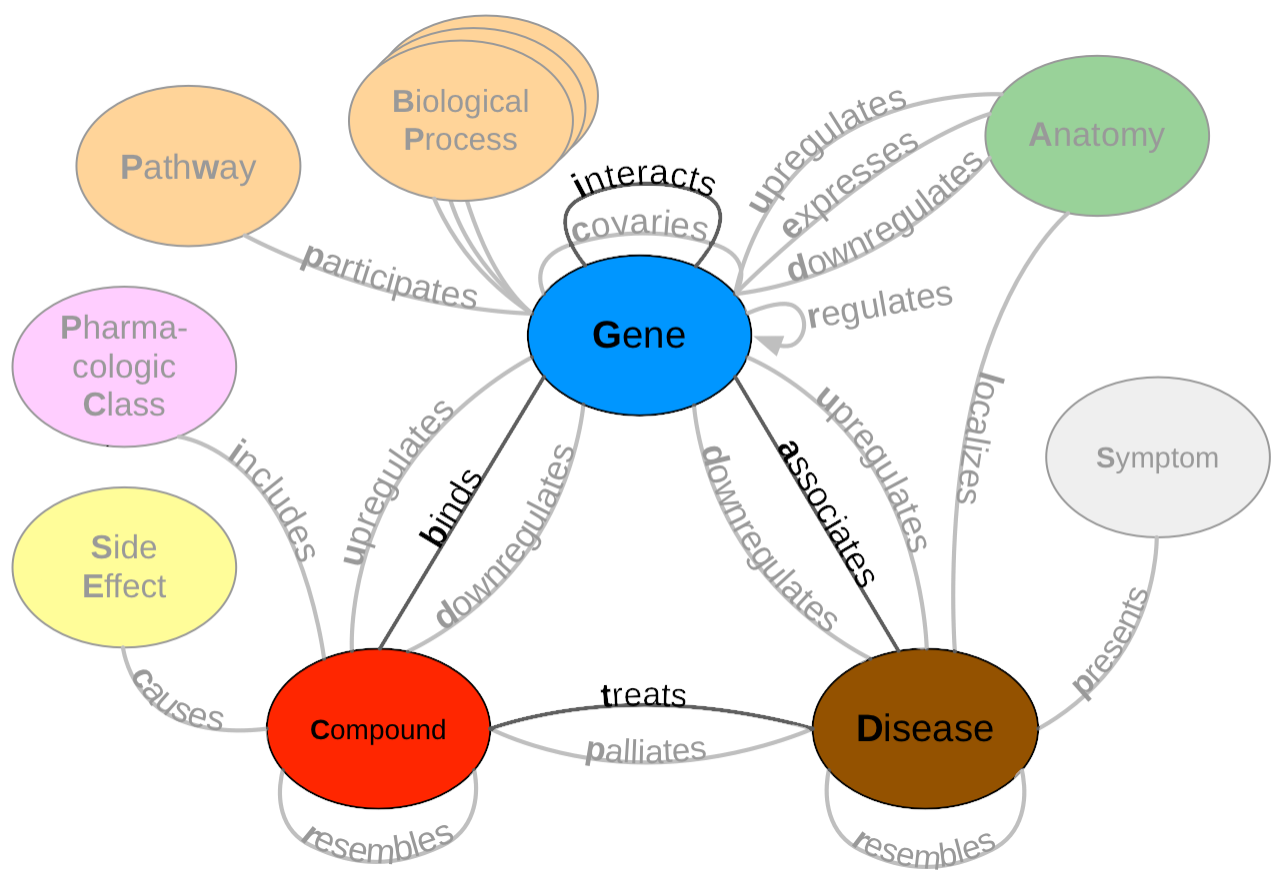
This work uses a subset of Hetionet v1 (bolded in the resource schema below), which is a heterogenous network that contains pharmacological and biological information in the form of nodes and edges. This network was made from publicly available data, which is usually populated via manual curation. Manual curation is time consuming and difficult to scale as the rate of publications continues to rise. A recently introduced "Data Programming" paradigm can circumvent this issue by being able to generate large annotated datasets quickly. This paradigm combines distant supervision with simple rules and heuristics written as labeling functions to automatically annotate large datasets. Unfortunately, it takes a significant amount of time and effort to write a useful label function. Because of this fact, we aimed to speed up this process by re-using label functions across edge types. Read the full paper here.
Directories
Described below are the main folders for this project. For convention the folder names are based on the schema shown above.
| Name | Descirption |
|---|---|
| compound_disease | Head folder that contains all relationships compounds and diseases may share |
| compound_gene | Head folder that contains all relationships compounds and genes may share |
| disease_gene | Head folder that contains all realtionships disease and genes may share |
| gene_gene | Head folder than contains all realtionships genes may share with each other |
| dependency cluster | This folder contains preprocessed results from the "A global network of biomedical relationships derived from text" paper. |
| figures | This folder contains figures for this work |
| modules | This folder contains helper scripts that this work uses |
| playground | This folder contains ancient code designed to test and understand the snorkel package. |
Installing/Setting Up The Conda Environment
Snorkeling uses conda as a python package manager. Before moving on to the instructions below, please make sure to have it installed. Download conda here!!
Once everything has been installed, type following command in the terminal:
conda env create --file environment.ymlYou can activate the environment by using the following command:
source activate snorkelingNote: If you want to leave the environment, just enter the following command:
source deactivate License
This repository is dual licensed as BSD 3-Clause and CC0 1.0, meaning any repository content can be used under either license. This licensing arrangement ensures source code is available under an OSI-approved License, while non-code content — such as figures, data, and documentation — is maximally reusable under a public domain dedication.