Spinal Cord Toolbox
Spinal Cord Toolbox (SCT) is a comprehensive, free and open-source set of command-line tools dedicated to the processing and analysis of spinal cord MRI data.
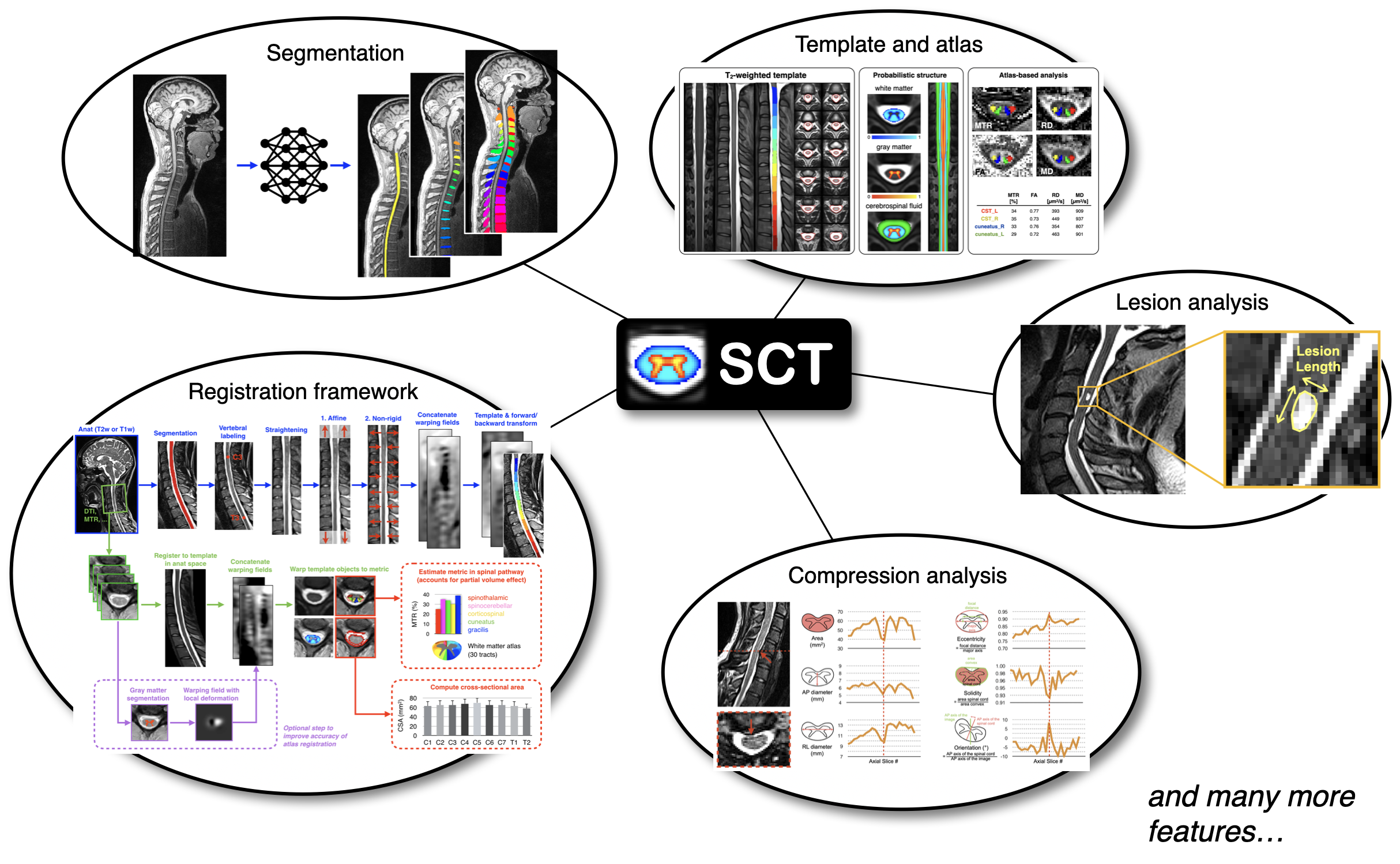
Key Features
- Segmentation of the spinal cord and gray matter
- Segmentation of pathologies (e.g. multiple sclerosis lesions)
- Detection of anatomical highlights (e.g. ponto-medullary junction, spinal cord centerline, vertebral levels)
- Registration to template, and deformation (e.g. straightening)
- Motion correction for diffusion and functional MRI time series
- Computation of quantitative MRI metrics (e.g. diffusion tensor imaging, magnetization transfer)
- Texture analysis (e.g. grey level co-occurrence matrix)
- Extraction of metrics within anatomical regions (e.g. white matter tracts)
- Manual labeling and segmentation via a Graphical User Interface (GUI)
- Warping field creation and application
- NIFTI volume manipulation tools for common operations
Installation
For macOS and Linux users, the simplest way to install SCT is to download the latest release, then launch the install script:
./install_sct
For more complex installation setups (Windows users, Docker, FSLeyes integration), see the Installation page.
Usage
Once installed, there are three main ways to use SCT:
1. Command-line tools
The primary way to invoke SCT is through terminal commands. For example:
$ sct_deepseg_sc -i t2.nii.gz -c t2
Cropping the image around the spinal cord...
Normalizing the intensity...
Segmenting the spinal cord using deep learning on 2D patches...
Reassembling the image...
Resampling the segmentation to the native image resolution using linear interpolation...
Binarizing the resampled segmentation...
Compute shape analysis: 100%|################| 55/55 [00:00<00:00, 106.05iter/s]
Done! To view results, type:
fsleyes t2.nii.gz -cm greyscale t2_seg.nii.gz -cm red -a 70.0 &For a full overview of the available commands, see the Command-Line Tools page.
2. Multi-command pipelines
To facilitate multi-subject analyses, commands can be chained together to build processing pipelines. The best starting point for constructing a typical pipeline is the batch_processing.sh script, which is provided with your installation of SCT.
3. GUI (FSLeyes integration)
SCT provides a graphical user interface via a FSLeyes plugin. For more details, see the FSLeyes Integration page.
Who is using SCT?
SCT is trusted by the research labs of many highly-regarded institutions worldwide. A full list of endorsements can be found on the Testimonials page.
For a list of neuroimaging studies that depend on SCT, visit the Studies using SCT page.
Contact
If you have any questions or concerns, and would like to get in touch with the developers of Spinal Cord Toolbox, the best way to do so is via the Spinalcordmri.org forum.
Otherwise, if you would like to stay up to date on SCT announcements (new releases, hands-on courses, etc.), please subscribe to the Spinalcordmri.org mailing list.
License
SCT is made available under the LGPLv3 license. For more details, see LICENSE.
Contributing
We happily welcome contributions. Please see the Contributing page of the developer Wiki for more information.
Warning
️Medical Disclaimer
All content found in the Spinal Cord Toolbox repository and spinalcordtoolbox.com website, including: text, images, audio, or other formats were created for informational purposes only. The content is not intended to be a substitute for professional medical advice, diagnosis, or treatment. Always seek the advice of your physician or other qualified health provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay in seeking it because of something you have read on this website.
If you think you may have a medical emergency, call your doctor, go to the emergency department, or call your local emergency number immediately. Spinal Cord Toolbox does not recommend or endorse any specific tests, physicians, products, procedures, opinions, or other information that may be mentioned on spinalcordtoolbox.com. Reliance on any information provided by spinalcordtoolbox.com, Spinal Cord Toolbox contributors, contracted writers, or medical professionals presenting content for publication to spinalcordtoolbox.com is solely at your own risk.