dcomtois / Summarytools
Programming Languages
Projects that are alternatives of or similar to Summarytools
summarytools 
The following vignettes complement this page:
Recommendations for Using summarytools With
Rmarkdown
Introduction to
summarytools
– Contents similar to this page (minus installation instructions), with
fancier table stylings.
1. Overview
summarytools is a an R package for data exploration and simple reporting.
Four functions are at its core:
| Function | Description |
|---|---|
freq() |
Frequency Tables featuring counts, proportions, as well as missing data information |
ctable() |
Cross-Tabulations (joint frequencies) between pairs of discrete variables featuring marginal sums as well as row, column or total proportions |
descr() |
Descriptive (Univariate) Statistics for numerical data featuring common measures of central tendency and dispersion |
dfSummary() |
Extensive Data Frame Summaries featuring type-specific information for all variables in a data frame: univariate statistics and/or frequency distributions, bar charts or histograms, as well as missing data counts. Very useful to quickly detect anomalies and identify trends at a glance |
1.1 Motivation
The package was developed with the following objectives in mind:
- Provide a coherent set of easy-to-use descriptive functions that are akin to those included in commercial statistical software suites such as SAS, SPSS, and Stata
- Offer flexibility in terms of output format & content
- Integrate well with commonly used software & tools for reporting (the RStudio IDE, Rmarkdown, and knitr) while also allowing for standalone, simple report generation from any R interface
On a more personal level, I simply wish to share with the R community and the scientific community at large the functions I first developed for myself, that I ultimately realized would benefit a lot of people who are looking for the same thing I was seeking in the first place.
Support summarytools’ Development With a Small Donation
Some package developers and maintainers get paid to do exactly that. They may also work in teams. This is not my case. Seeing the package grow in popularity was and still is in itself a rewarding experience, but I won’t lie; keeping up with the maintenance, feature requests and other features I have in mind takes more time than I can afford.
So if you find summarytools useful and want to support its development, please consider making a small donation using the PayPal button. In exchange, on top of contributing to the package and helping out other data scientists, students and researchers, you’ll get:
- My sincere gratitude
- Your name listed in the Sponsors section of this page
- My personal commitment to dedicate more time to the package’s development
1.2 Redirecting Outputs
Results can be
- Displayed in the R console as plain text
- Rendered as html and shown in a Web browser or in RStudio’s Viewer pane
- Written to, or appended to plain text, markdown, or html files
1.3 Other Characteristics
- Pipe-Friendly:
-
Multilingual:
- Built-in translations exist for French, Portuguese, Spanish, Russian and Turkish
- Users can easily add custom translations or modify existing sets of translations as needed
-
Weights-enabled: except for
dfSummary(), all core functions support sampling weights -
Flexible:
- Default values for most function arguments can be modified using
st_options(); this simplifies coding and minimizes redundancy - Pander options can be used for text / markdown tables
- Base R’s
format()parameters are supported; this is especially useful to set thousands separators, among several other possibilities - Bootstrap CSS used by default with html outputs, and user-defined classes can be added at will
- Default values for most function arguments can be modified using
1.4 Installing summarytools
Required Software
Additional software is used by summarytools to fine-tune graphics as well as offer interactive features. If installing summarytools for the first time, click on the link corresponding to your Operating System to get detailed instructions. Note that on Windows, no additional software is required.
Mac OS X
Ubuntu / Debian / Mint
Older Ubuntu (14 and 16)
Fedora / Red Hat / CentOS
Solaris
Installing From GitHub
This is the recommended method, as some minor fixes and improvements are regularly added.
install.packages("remotes") # Using devtools is also possible
library(remotes)
install_github("rapporter/pander") # Necessary for optimal results!
install_github("dcomtois/summarytools")
Installing From CRAN
CRAN versions are stable but are not updated as often as the GitHub versions.
install.packages("summarytools")
1.5 Latest Features (versions 0.9.7 and 0.9.8)
- Performance and formatting improvements
- The
stview()function which ensures the package’s ownview()method
is used (avoiding potential conflicts with other packages’ versions of that method) - Several other features (see NEWS.md or try
news(package="summarytools"))
2. The Four Core Functions
2.1 Frequency Tables With freq()
The freq() function generates frequency tables with counts,
proportions, as well as missing data information.
freq(iris$Species, plain.ascii = FALSE, style = "rmarkdown")
Frequencies
iris$Species
Type: Factor
| Freq | % Valid | % Valid Cum. | % Total | % Total Cum. | |
|---|---|---|---|---|---|
| setosa | 50 | 33.33 | 33.33 | 33.33 | 33.33 |
| versicolor | 50 | 33.33 | 66.67 | 33.33 | 66.67 |
| virginica | 50 | 33.33 | 100.00 | 33.33 | 100.00 |
| <NA> | 0 | 0.00 | 100.00 | ||
| Total | 150 | 100.00 | 100.00 | 100.00 | 100.00 |
In this first example, the plain.ascii and style arguments were
specified. However, since we have defined them globally for this
document using st_options(), they are redundant and will be omitted
from hereon.
2.1.1 Formatting Numbers With format()’s Arguments
As of version 0.9.8, it is possible to use base R’s
format()
parameters when calling freq() or any other core function. Some of the
most useful are big.mark, which inserts thousands separators, and
decimal.mark, which allows using commas instead of dots as decimal
separator (useful in several locales). Note that decimal marks can also
be set globally with the R option OutDec (e.g.
options(OutDec = ",")). The formatting is applied in the heading
section as well as in the results tables:
set.seed(2835)
Random_numbers <- sample(c(5e3, 5e4, 5e5), size = 1e4, replace = TRUE, prob = c(.12, .36, .52))
freq(Random_numbers, big.mark = ",", cumul = FALSE, headings = FALSE)
## setting plain.ascii to FALSE
| Freq | % Valid | % Total | |
|---|---|---|---|
| 5,000 | 1,237 | 12.37 | 12.37 |
| 50,000 | 3,605 | 36.05 | 36.05 |
| 500,000 | 5,158 | 51.58 | 51.58 |
| <NA> | 0 | 0.00 | |
| Total | 10,000 | 100.00 | 100.00 |
# We can also use format() arguments with print / view
print(freq(Random_numbers, cumul = FALSE, headings = FALSE), big.mark = " ", decimal.mark = ".")
## setting plain.ascii to FALSE
| Freq | % Valid | % Total | |
|---|---|---|---|
| 5 000 | 1 237 | 12.37 | 12.37 |
| 50 000 | 3 605 | 36.05 | 36.05 |
| 500 000 | 5 158 | 51.58 | 51.58 |
| <NA> | 0 | 0.00 | |
| Total | 10 000 | 100.00 | 100.00 |
2.1.2 Ignoring Missing Data
The report.nas argument can be set to FALSE in order to ignore
missing values (NA’s). Doing so has the following effects on the
resulting table:
- The <NA> row is omitted
- The % Total and % Total Cum. (cumulative) columns are also omitted
- The % Valid column simply becomes %
- The % Valid Cum. column simply becomes % Cum.
freq(iris$Species, report.nas = FALSE, headings = FALSE)
## setting plain.ascii to FALSE
| Freq | % | % Cum. | |
|---|---|---|---|
| setosa | 50 | 33.33 | 33.33 |
| versicolor | 50 | 33.33 | 66.67 |
| virginica | 50 | 33.33 | 100.00 |
| Total | 150 | 100.00 | 100.00 |
Note that the headings = FALSE parameter suppresses the heading
section. (The heading section consists of a title, as well as various
metadata elements: object names, labels, by-groups, and so on.
2.1.3 Minimal Frequency Tables
By “switching off” all optional elements, a much simpler table will be produced:
freq(iris$Species, report.nas = FALSE, totals = FALSE,
cumul = FALSE, headings = FALSE)
## setting plain.ascii to FALSE
| Freq | % | |
|---|---|---|
| setosa | 50 | 33.33 |
| versicolor | 50 | 33.33 |
| virginica | 50 | 33.33 |
2.1.4 Multiple Frequency Tables
To generate frequency tables for all variables in a data frame, one
could use lapply(). However, this is not required since freq()
handles whole data frames, too:
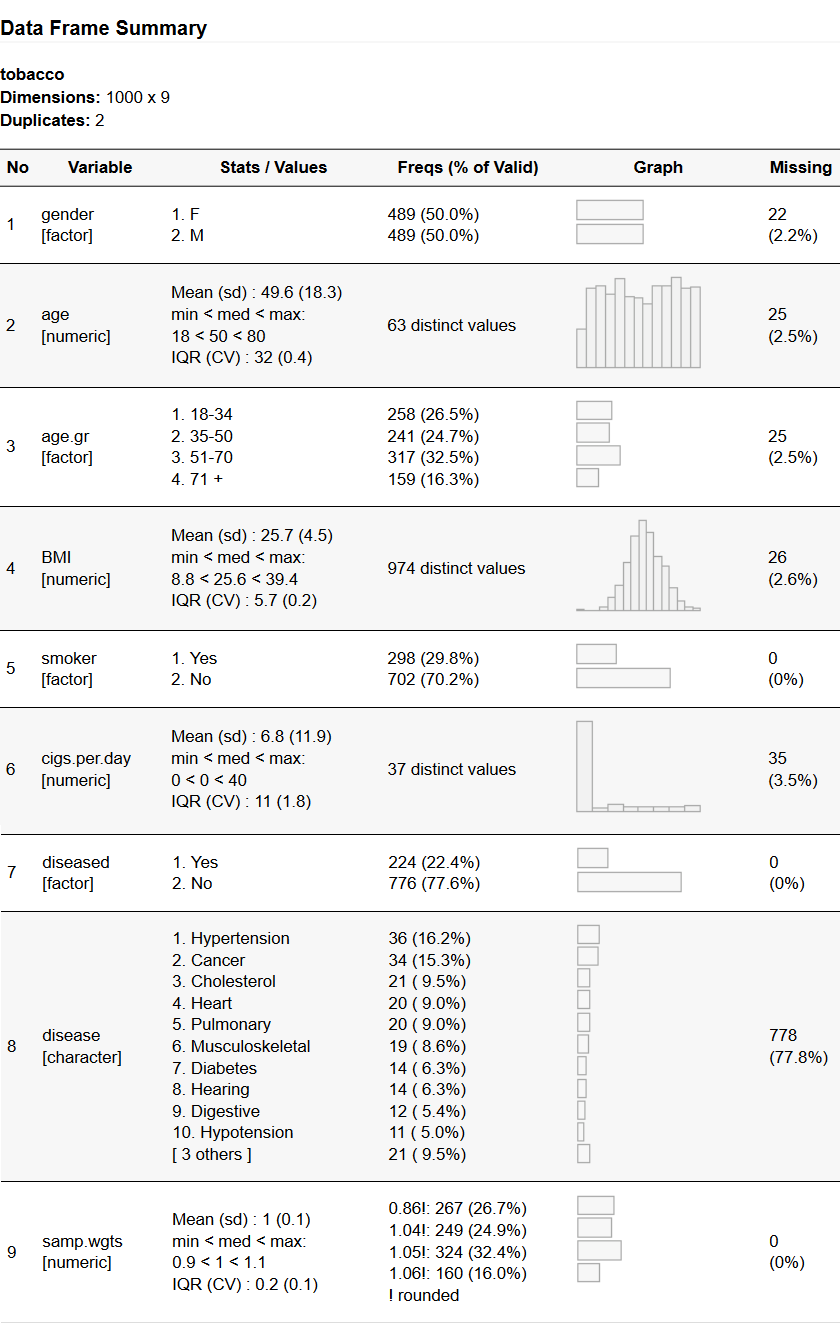
freq(tobacco)
To avoid cluttering the results, numerical columns having more than 25
distinct values are ignored. This threshold of 25 can be changed by
using st_options(); for example, to change it to 10, we’d use
st_options(freq.ignore.threshold = 10).
Note: the tobacco data frame contains simulated data and is included in the package. Another simulated data frame is included: exams. Both have French versions (tabagisme, examens).
2.1.5 Subsetting (Filtering) Frequency Tables
The rows parameter allows subsetting frequency tables; we can use this
parameter in different ways:
- To filter rows by their order of appearance, we use a numerical
vector;
rows = 1:10will show the frequencies for the first 10 values only - To filter rows by name, we can use
- a character vector specifying the exact row names we wish to keep in the results
- a single character string which will be used as a regular
expression to select the matching column(s); see
?regexfor more information on regular expressions
Used in combination with the order argument, the subsetting feature
can be quite practical. For a character variable containing a large
number of distinct values, showing only the most frequent is easily
done:
freq(tobacco$disease, order = "freq", rows = 1:5, headings = FALSE)
## setting plain.ascii to FALSE
| Freq | % Valid | % Valid Cum. | % Total | % Total Cum. | |
|---|---|---|---|---|---|
| Hypertension | 36 | 16.22 | 16.22 | 3.60 | 3.60 |
| Cancer | 34 | 15.32 | 31.53 | 3.40 | 7.00 |
| Cholesterol | 21 | 9.46 | 40.99 | 2.10 | 9.10 |
| Heart | 20 | 9.01 | 50.00 | 2.00 | 11.10 |
| Pulmonary | 20 | 9.01 | 59.01 | 2.00 | 13.10 |
| (Other) | 91 | 40.99 | 100.00 | 9.10 | 22.20 |
| <NA> | 778 | 77.80 | 100.00 | ||
| Total | 1000 | 100.00 | 100.00 | 100.00 | 100.00 |
Instead of "freq", we can use "-freq" to reverse the ordering and
get results ranked from lowest to highest in frequency.
To account for the frequencies of unshown values, the “(Other)” row is automatically added.
2.1.6 Collapsible Sections
When generating html results, use the collapse = TRUE argument with
print() or view() to get collapsible sections; clicking on the
variable name in the heading section will collapse / reveal the
frequency table (results not shown).
view(freq(tobacco), collapse = TRUE)
2.2 Cross-Tabulations with ctable()
ctable() generates cross-tabulations (joint frequencies) for pairs of
categorical variables.
Since markdown does not support multiline table headings (but does accept html code), we’ll use the html rendering feature for this section.
Using the tobacco data frame, we’ll cross-tabulate the two categorical variables smoker and diseased.
print(ctable(x = tobacco$smoker, y = tobacco$diseased, prop = "r"),
method = "render")
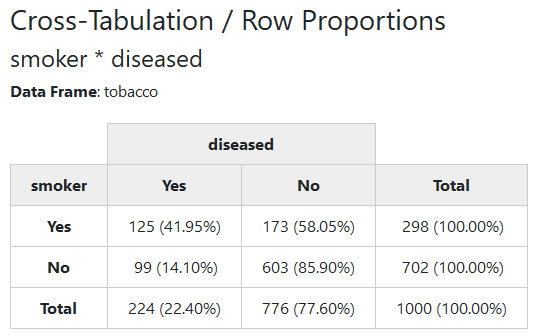
2.2.1 Row, Column or Total Proportions
Row proportions are shown by default. To display column or total
proportions, use prop = "c" or prop = "t", respectively. To omit
proportions altogether, use prop = "n".
2.2.2 Minimal Cross-Tabulations
By “switching off” all optional features, we get a simple “2 x 2” table:
with(tobacco,
print(ctable(x = smoker, y = diseased, prop = 'n',
totals = FALSE, headings = FALSE),
method = "render"))
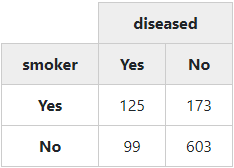
2.2.3 Chi-Square (𝛘2), Odds Ratio and Risk Ratio
To display the chi-square statistic, set chisq = TRUE. For 2 x 2
tables, use OR and RR to show odds ratio and risk ratio (also called
relative risk), respectively. Those can be set to TRUE, in which case
95% confidence intervals will be shown; to use alternate confidence
levels, use for example OR = .90.
To show how pipes can be used with summarytools, we’ll use
magrittr’s %$% and %>% operators:
library(magrittr)
tobacco %$% # Acts like with(tobacco, ...)
ctable(smoker, diseased,
chisq = TRUE, OR = TRUE, RR = TRUE,
headings = FALSE) %>%
print(method = "render")
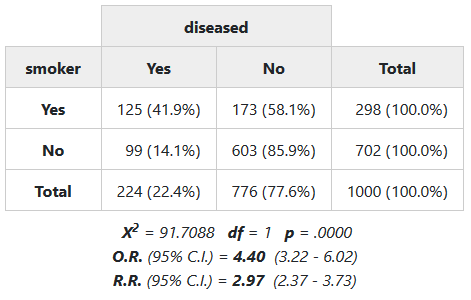
2.3 Descriptive Statistics With descr()
descr() generates descriptive / univariate statistics, i.e. common
central tendency statistics and measures of dispersion. It accepts
single vectors as well as data frames; in the latter case, all
non-numerical columns are ignored, with a message to that effect.
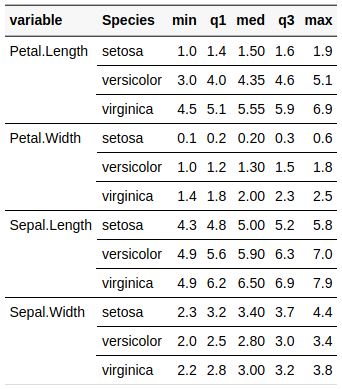
descr(iris)
Descriptive Statistics
iris
N: 150
| Petal.Length | Petal.Width | Sepal.Length | Sepal.Width | |
|---|---|---|---|---|
| Mean | 3.76 | 1.20 | 5.84 | 3.06 |
| Std.Dev | 1.77 | 0.76 | 0.83 | 0.44 |
| Min | 1.00 | 0.10 | 4.30 | 2.00 |
| Q1 | 1.60 | 0.30 | 5.10 | 2.80 |
| Median | 4.35 | 1.30 | 5.80 | 3.00 |
| Q3 | 5.10 | 1.80 | 6.40 | 3.30 |
| Max | 6.90 | 2.50 | 7.90 | 4.40 |
| MAD | 1.85 | 1.04 | 1.04 | 0.44 |
| IQR | 3.50 | 1.50 | 1.30 | 0.50 |
| CV | 0.47 | 0.64 | 0.14 | 0.14 |
| Skewness | -0.27 | -0.10 | 0.31 | 0.31 |
| SE.Skewness | 0.20 | 0.20 | 0.20 | 0.20 |
| Kurtosis | -1.42 | -1.36 | -0.61 | 0.14 |
| N.Valid | 150.00 | 150.00 | 150.00 | 150.00 |
| Pct.Valid | 100.00 | 100.00 | 100.00 | 100.00 |
2.3.1 Transposing and Selecting Statistics
Results can be transposed by using transpose = TRUE, and statistics
can be selected using the stats argument:
descr(iris, stats = c("mean", "sd"), transpose = TRUE, headings = FALSE)
| Mean | Std.Dev | |
|---|---|---|
| Petal.Length | 3.76 | 1.77 |
| Petal.Width | 1.20 | 0.76 |
| Sepal.Length | 5.84 | 0.83 |
| Sepal.Width | 3.06 | 0.44 |
See ?descr for a list of all available statistics. Special values
“all”, “fivenum”, and “common” are also valid values for the stats
argument. The default value is “all”.
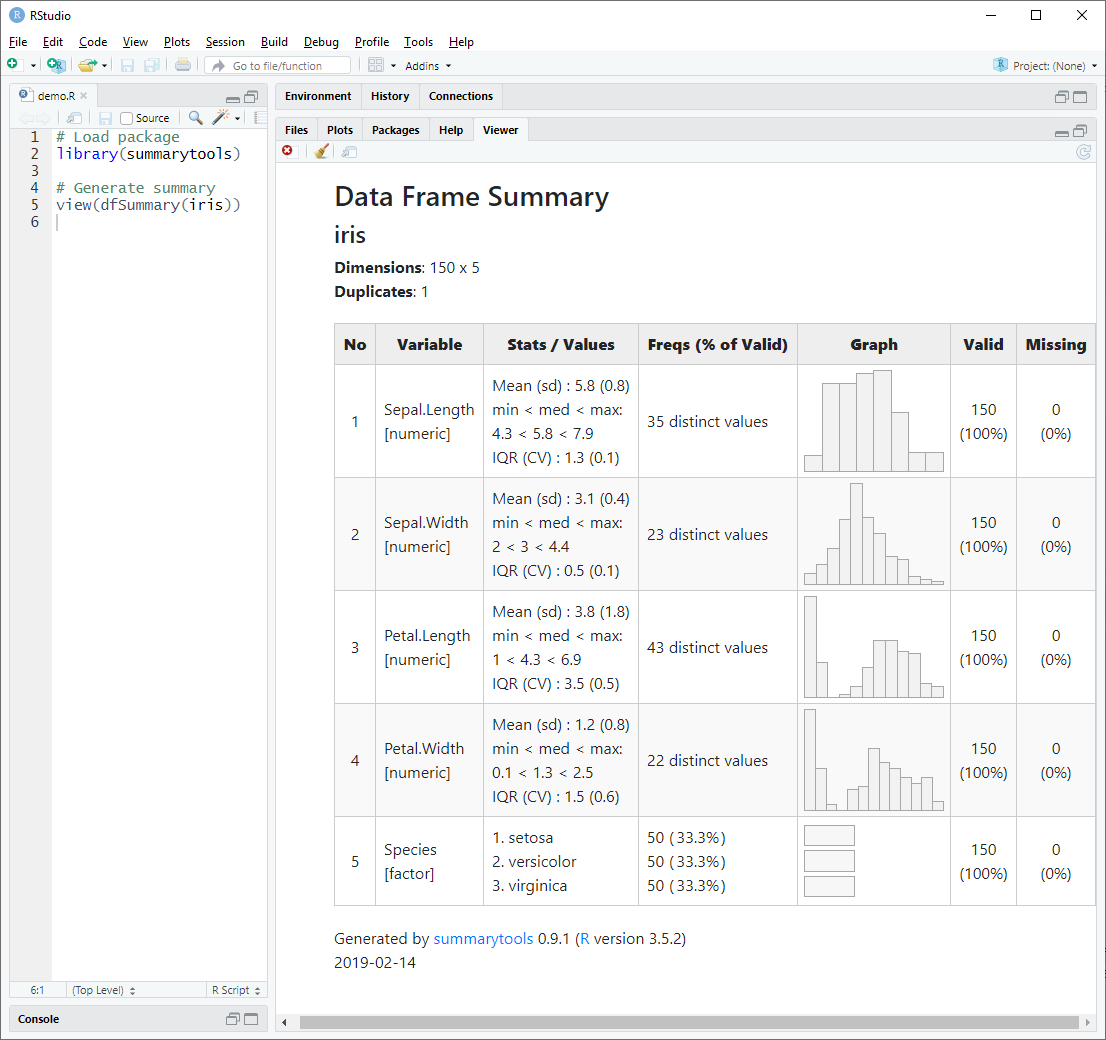
2.4 Data Frame Summaries With dfSummary()
dfSummary() creates a summary table with statistics, frequencies and
graphs for all variables in a data frame. The information displayed is
type-specific (character, factor, numeric, date) and also varies
according to the number of distinct values.
To see the results in RStudio’s Viewer (or in the default Web browser if
working in another IDE or from a terminal window), we use the view()
function:
view(dfSummary(iris))
2.4.1 Using dfSummary() in Rmarkdown Documents
When using dfSummary() in Rmarkdown documents, it is generally a
good idea to exclude a column or two to avoid margin overflow. Since the
Valid and Missing columns are redundant, we can drop either one of
them.
dfSummary(tobacco, plain.ascii = FALSE, style = "grid",
graph.magnif = 0.75, valid.col = FALSE, tmp.img.dir = "/tmp")
The tmp.img.dir parameter is mandatory when generating
dfSummaries in Rmarkdown documents, except for html rendering. The
explanation for this can be found further below.
2.4.2 Advanced Features
The dfSummary() function also
- Reports the number of duplicate records in the heading section
- Detects UPC/EAN codes (barcode numbers) and doesn’t calculate irrelevant statistics for them
- Detects email addresses and reports counts of valid, invalid and duplicate addresses
2.4.3 Excluding Columns
Although most columns can be excluded using the function’s parameters, it is also possible to delete them with the following syntax (results not shown):
dfs <- dfSummary(iris)
dfs$Variable <- NULL # This deletes the "Variable" column
3. Grouped Statistics Using stby()
To produce optimal results, summarytools has its own version of the
base by() function. It’s called stby(), and we use it exactly as we
would by():
(iris_stats_by_species <- stby(data = iris,
INDICES = iris$Species,
FUN = descr, stats = "common", transpose = TRUE))
## Non-numerical variable(s) ignored: Species
Descriptive Statistics
iris
Group: Species = setosa
N: 50
| Mean | Std.Dev | Min | Median | Max | N.Valid | Pct.Valid | |
|---|---|---|---|---|---|---|---|
| Petal.Length | 1.46 | 0.17 | 1.00 | 1.50 | 1.90 | 50.00 | 100.00 |
| Petal.Width | 0.25 | 0.11 | 0.10 | 0.20 | 0.60 | 50.00 | 100.00 |
| Sepal.Length | 5.01 | 0.35 | 4.30 | 5.00 | 5.80 | 50.00 | 100.00 |
| Sepal.Width | 3.43 | 0.38 | 2.30 | 3.40 | 4.40 | 50.00 | 100.00 |
Group: Species = versicolor
N: 50
| Mean | Std.Dev | Min | Median | Max | N.Valid | Pct.Valid | |
|---|---|---|---|---|---|---|---|
| Petal.Length | 4.26 | 0.47 | 3.00 | 4.35 | 5.10 | 50.00 | 100.00 |
| Petal.Width | 1.33 | 0.20 | 1.00 | 1.30 | 1.80 | 50.00 | 100.00 |
| Sepal.Length | 5.94 | 0.52 | 4.90 | 5.90 | 7.00 | 50.00 | 100.00 |
| Sepal.Width | 2.77 | 0.31 | 2.00 | 2.80 | 3.40 | 50.00 | 100.00 |
Group: Species = virginica
N: 50
| Mean | Std.Dev | Min | Median | Max | N.Valid | Pct.Valid | |
|---|---|---|---|---|---|---|---|
| Petal.Length | 5.55 | 0.55 | 4.50 | 5.55 | 6.90 | 50.00 | 100.00 |
| Petal.Width | 2.03 | 0.27 | 1.40 | 2.00 | 2.50 | 50.00 | 100.00 |
| Sepal.Length | 6.59 | 0.64 | 4.90 | 6.50 | 7.90 | 50.00 | 100.00 |
| Sepal.Width | 2.97 | 0.32 | 2.20 | 3.00 | 3.80 | 50.00 | 100.00 |
3.1 Special Case of descr() with stby()
When used to produce split-group statistics for a single variable,
stby() assembles everything into a single table instead of displaying
a series of one-column tables.
with(tobacco, stby(data = BMI, INDICES = age.gr,
FUN = descr, stats = c("mean", "sd", "min", "med", "max")))
Descriptive Statistics
BMI by age.gr
Data Frame: tobacco
N: 258
| 18-34 | 35-50 | 51-70 | 71 + | |
|---|---|---|---|---|
| Mean | 23.84 | 25.11 | 26.91 | 27.45 |
| Std.Dev | 4.23 | 4.34 | 4.26 | 4.37 |
| Min | 8.83 | 10.35 | 9.01 | 16.36 |
| Median | 24.04 | 25.11 | 26.77 | 27.52 |
| Max | 34.84 | 39.44 | 39.21 | 38.37 |
3.2 Using stby() With ctable()
The syntax is a little trickier for this combination, so here is an example (results not shown):
stby(list(x = tobacco$smoker, y = tobacco$diseased),
INDICES = tobacco$gender, FUN = ctable)
# or equivalently
with(tobacco,
stby(list(x = smoker, y = diseased),
INDICES = gender, FUN = ctable))
4. Grouped Statistics Using dplyr::group_by()
To create grouped statistics with freq(), descr() or dfSummary(),
it is possible to use dplyr’s group_by() as an alternative to
stby(). Syntactic differences aside, one key distinction is that
group_by() considers NA values on the grouping variable(s) as a
valid category, albeit with a warning message suggesting the use of
forcats::fct_explicit_na to make NA’s explicit in factors. Following
this advice, we get:
library(dplyr)
tobacco$gender %<>% forcats::fct_explicit_na()
tobacco %>% group_by(gender) %>% descr(stats = "fivenum")
## Non-numerical variable(s) ignored: age.gr, smoker, diseased, disease
Descriptive Statistics
tobacco
Group: gender = F
N: 489
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 18.00 | 9.01 | 0.00 | 0.86 |
| Q1 | 34.00 | 22.98 | 0.00 | 0.86 |
| Median | 50.00 | 25.87 | 0.00 | 1.04 |
| Q3 | 66.00 | 29.48 | 10.50 | 1.05 |
| Max | 80.00 | 39.44 | 40.00 | 1.06 |
Group: gender = M
N: 489
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 18.00 | 8.83 | 0.00 | 0.86 |
| Q1 | 34.00 | 22.52 | 0.00 | 0.86 |
| Median | 49.50 | 25.14 | 0.00 | 1.04 |
| Q3 | 66.00 | 27.96 | 11.00 | 1.05 |
| Max | 80.00 | 36.76 | 40.00 | 1.06 |
Group: gender = (Missing)
N: 22
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 19.00 | 20.24 | 0.00 | 0.86 |
| Q1 | 36.00 | 24.97 | 0.00 | 1.04 |
| Median | 55.50 | 27.16 | 0.00 | 1.05 |
| Q3 | 64.00 | 30.23 | 10.00 | 1.05 |
| Max | 80.00 | 32.43 | 28.00 | 1.06 |
5. Creating Tidy Tables With tb()
When generating freq() or descr() tables, it is possible to turn the
results into “tidy” tables with the use of the tb() function (think of
tb as a diminutive for tibble). For example:
library(magrittr)
iris %>% descr(stats = "common") %>% tb()
## # A tibble: 4 x 8
## variable mean sd min med max n.valid pct.valid
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Petal.Length 3.76 1.77 1 4.35 6.9 150 100
## 2 Petal.Width 1.20 0.762 0.1 1.3 2.5 150 100
## 3 Sepal.Length 5.84 0.828 4.3 5.8 7.9 150 100
## 4 Sepal.Width 3.06 0.436 2 3 4.4 150 100
iris$Species %>% freq(cumul = FALSE, report.nas = FALSE) %>% tb()
## setting plain.ascii to FALSE
## # A tibble: 3 x 3
## Species freq pct
## <fct> <dbl> <dbl>
## 1 setosa 50 33.3
## 2 versicolor 50 33.3
## 3 virginica 50 33.3
By definition, no total rows are part of tidy tables, and the row names are converted to a regular column. Note that for displaying tibbles using Rmarkdown, the knitr chunk option ‘results’ should be set to “markup” instead of “asis”.
5.1 Tidy Split-Group Statistics
Here are some examples showing how lists created using stby() or
group_by() can be transformed into tidy tibbles.
grouped_descr <- stby(data = exams, INDICES = exams$gender,
FUN = descr, stats = "common")
grouped_descr %>% tb()
## # A tibble: 12 x 9
## gender variable mean sd min med max n.valid pct.valid
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Girl economics 72.5 7.79 62.3 70.2 89.6 14 93.3
## 2 Girl english 73.9 9.41 58.3 71.8 93.1 14 93.3
## 3 Girl french 71.1 12.4 44.8 68.4 93.7 14 93.3
## 4 Girl geography 67.3 8.26 50.4 67.3 78.9 15 100
## 5 Girl history 71.2 9.17 53.9 72.9 86.4 15 100
## 6 Girl math 73.8 9.03 55.6 74.8 86.3 14 93.3
## 7 Boy economics 75.2 9.40 60.5 71.7 94.2 15 100
## 8 Boy english 77.8 5.94 69.6 77.6 90.2 15 100
## 9 Boy french 76.6 8.63 63.2 74.8 94.7 15 100
## 10 Boy geography 73 12.4 47.2 71.2 96.3 14 93.3
## 11 Boy history 74.4 11.2 54.4 72.6 93.5 15 100
## 12 Boy math 73.3 9.68 60.5 72.2 93.2 14 93.3
The order parameter controls row ordering:
grouped_descr %>% tb(order = 2)
## # A tibble: 12 x 9
## gender variable mean sd min med max n.valid pct.valid
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Girl economics 72.5 7.79 62.3 70.2 89.6 14 93.3
## 2 Boy economics 75.2 9.40 60.5 71.7 94.2 15 100
## 3 Girl english 73.9 9.41 58.3 71.8 93.1 14 93.3
## 4 Boy english 77.8 5.94 69.6 77.6 90.2 15 100
## 5 Girl french 71.1 12.4 44.8 68.4 93.7 14 93.3
## 6 Boy french 76.6 8.63 63.2 74.8 94.7 15 100
## 7 Girl geography 67.3 8.26 50.4 67.3 78.9 15 100
## 8 Boy geography 73 12.4 47.2 71.2 96.3 14 93.3
## 9 Girl history 71.2 9.17 53.9 72.9 86.4 15 100
## 10 Boy history 74.4 11.2 54.4 72.6 93.5 15 100
## 11 Girl math 73.8 9.03 55.6 74.8 86.3 14 93.3
## 12 Boy math 73.3 9.68 60.5 72.2 93.2 14 93.3
Setting order = 3 changes the order of the sort variables exactly as
with order = 2, but it also reorders the columns:
grouped_descr %>% tb(order = 3)
## # A tibble: 12 x 9
## variable gender mean sd min med max n.valid pct.valid
## <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 economics Girl 72.5 7.79 62.3 70.2 89.6 14 93.3
## 2 economics Boy 75.2 9.40 60.5 71.7 94.2 15 100
## 3 english Girl 73.9 9.41 58.3 71.8 93.1 14 93.3
## 4 english Boy 77.8 5.94 69.6 77.6 90.2 15 100
## 5 french Girl 71.1 12.4 44.8 68.4 93.7 14 93.3
## 6 french Boy 76.6 8.63 63.2 74.8 94.7 15 100
## 7 geography Girl 67.3 8.26 50.4 67.3 78.9 15 100
## 8 geography Boy 73 12.4 47.2 71.2 96.3 14 93.3
## 9 history Girl 71.2 9.17 53.9 72.9 86.4 15 100
## 10 history Boy 74.4 11.2 54.4 72.6 93.5 15 100
## 11 math Girl 73.8 9.03 55.6 74.8 86.3 14 93.3
## 12 math Boy 73.3 9.68 60.5 72.2 93.2 14 93.3
For more details, see ?tb.
5.2 A Bridge to Other Packages
summarytools objects are not always compatible with packages focused
on table formatting, such as
formattable or
kableExtra. However,
tb() can be used as a “bridge”, an intermediary step turning freq()
and descr() objects into simple tables that any package can work with.
Here is an example using kableExtra:
library(kableExtra)
library(magrittr)
stby(iris, iris$Species, descr, stats = "fivenum") %>%
tb(order = 3) %>%
kable(format = "html", digits = 2) %>%
collapse_rows(columns = 1, valign = "top")
6. Redirecting Output to Files
Using the file argument with print() or view(), we can write
outputs to a file, be it html, Rmd, md, or just plain text
(txt). The file extension is used to determine the type of content to
write out.
view(iris_stats_by_species, file = "~/iris_stats_by_species.html")
view(iris_stats_by_species, file = "~/iris_stats_by_species.md")
A Note About PDF documents
There is no direct way to create a PDF file with summarytools. One
option is to generate an html file and convert it to PDF using
Pandoc or
WK<html>TOpdf (the latter
gives better results than Pandoc with dfSummary() output). Another
option is to create an Rmd document using PDF as the output format,
but with a caveat: displaying graphs with dfSummary() will cause
vertical misalignment (we hope to resolve this issue in a future
version).
6.1 Appending Output Files
The append argument allows adding content to existing files generated
by summarytools. This is useful if we wish to include several
statistical tables in a single file. It is a quick alternative to
creating an Rmd document.
7. Global options
The following options can be set with st_options():
7.1 General Options
| Option name | Default | Note |
|---|---|---|
| style (1) | “simple” | Set to “rmarkdown” in .Rmd documents |
| plain.ascii | TRUE | Set to FALSE in .Rmd documents |
| round.digits | 2 | Number of decimals to show |
| headings | TRUE | Formerly “omit.headings” |
| footnote | “default” | Customize or set to NA to omit |
| display.labels | TRUE | Show variable / data frame labels in headings |
| bootstrap.css (2) | TRUE | Include Bootstrap 4 CSS in html output files |
| custom.css | NA | Path to your own CSS file |
| escape.pipe | FALSE | Useful for some Pandoc conversions |
| char.split (3) | 12 | Threshold for line-wrapping in column headings |
| subtitle.emphasis | TRUE | Controls headings formatting |
| lang | “en” | Language (always 2-letter, lowercase) |
- Applies to
freq(),ctable()anddescr();dfSummary()has its own style option (see section 7.2) - Set to FALSE in Shiny apps
- Affects only html outputs for
descr()andctable()
7.2 Function-Specific Options
| Option name | Default | Note |
|---|---|---|
| freq.cumul | TRUE | Display cumulative proportions in freq() |
| freq.totals | TRUE | Display totals row in freq() |
| freq.report.nas | TRUE | Display row and “valid” columns |
| freq.ignore.threshold (1) | 25 | Used to determine which vars to ignore |
| freq.silent | FALSE | Hide console messages |
| ctable.prop | “r” | Display row proportions by default |
| ctable.totals | TRUE | Show marginal totals |
| descr.stats | “all” | “fivenum”, “common” or vector of stats |
| descr.transpose | FALSE | Display stats in columns instead of rows |
| descr.silent | FALSE | Hide console messages |
| dfSummary.style | “multiline” | Can be set to “grid” as an alternative |
| dfSummary.varnumbers | TRUE | Show variable numbers in 1st col. |
| dfSummary.labels.col | TRUE | Show variable labels when present |
| dfSummary.graph.col | TRUE | Show graphs |
| dfSummary.valid.col | TRUE | Include the Valid column in the output |
| dfSummary.na.col | TRUE | Include the Missing column in the output |
| dfSummary.graph.magnif | 1 | Zoom factor for bar plots and histograms |
| dfSummary.silent | FALSE | Hide console messages |
| tmp.img.dir (2) | NA | Directory to store temporary images |
| use.x11 (3) | TRUE | Allow creation of Base64-encoded graphs |
- See section 2.1.4 for details
- Applies to
dfSummary()only - Set to FALSE in text-only environments
Examples
st_options() # Display all global options values
st_options('round.digits') # Display the value of a specific option
st_options(style = 'rmarkdown', # Set the value of one or several options
footnote = NA) # Turn off the footnote for all html output
8. Overriding Formatting Attributes
When a summarytools object is created, its formatting attributes are
stored within it. However, we can override most of them when using
print() or view().
8.1 Overriding Function-Specific Arguments
This table indicates what arguments can be used with print() or
view() to override formatting attributes. Base R’s format() function
arguments also apply, even though they are not reproduced here.
| Argument | freq | ctable | descr | dfSummary |
|---|---|---|---|---|
| style | x | x | x | x |
| round.digits | x | x | x | |
| plain.ascii | x | x | x | x |
| justify | x | x | x | x |
| headings | x | x | x | x |
| display.labels | x | x | x | x |
| varnumbers | x | |||
| labels.col | x | |||
| graph.col | x | |||
| valid.col | x | |||
| na.col | x | |||
| col.widths | x | |||
| totals | x | x | ||
| report.nas | x | |||
| display.type | x | |||
| missing | x | |||
| split.tables (*) | x | x | x | x |
| caption (*) | x | x | x | x |
(*) These are pander options
8.2 Overriding Heading Contents
To change the information shown in the heading section, use the
following arguments with print() or view():
| Argument | freq | ctable | descr | dfSummary |
|---|---|---|---|---|
| Data.frame | x | x | x | x |
| Data.frame.label | x | x | x | x |
| Variable | x | x | x | |
| Variable.label | x | x | x | |
| Group | x | x | x | x |
| date | x | x | x | x |
| Weights | x | x | ||
| Data.type | x | |||
| Row.variable | x | |||
| Col.variable | x |
Example
In the following example, we will create and display a freq() object,
and then display it again, this time overriding three of its formatting
attributes, as well as one heading attribute.
(age_stats <- freq(tobacco$age.gr))
## setting plain.ascii to FALSE
Frequencies
tobacco$age.gr
Type: Factor
| Freq | % Valid | % Valid Cum. | % Total | % Total Cum. | |
|---|---|---|---|---|---|
| 18-34 | 258 | 26.46 | 26.46 | 25.80 | 25.80 |
| 35-50 | 241 | 24.72 | 51.18 | 24.10 | 49.90 |
| 51-70 | 317 | 32.51 | 83.69 | 31.70 | 81.60 |
| 71 + | 159 | 16.31 | 100.00 | 15.90 | 97.50 |
| <NA> | 25 | 2.50 | 100.00 | ||
| Total | 1000 | 100.00 | 100.00 | 100.00 | 100.00 |
print(age_stats, report.nas = FALSE, totals = FALSE, display.type = FALSE,
Variable.label = "Age Group")
Frequencies
tobacco$age.gr
Label: Age Group
| Freq | % | % Cum. | |
|---|---|---|---|
| 18-34 | 258 | 26.46 | 26.46 |
| 35-50 | 241 | 24.72 | 51.18 |
| 51-70 | 317 | 32.51 | 83.69 |
| 71 + | 159 | 16.31 | 100.00 |
8.3 Order of Priority for Parameters / Options
-
print()orview()parameters have precedence (overriding feature) -
freq() / ctable() / descr() / dfSummary()parameters come second - Global options set with
st_options()come third and act as default
9. Fine-Tuning Looks with CSS
When creating html reports, both Bootstrap’s CSS and summarytools.css are included by default. For greater control on the looks of html content, it is also possible to add class definitions in a custom CSS file.
Example
We need to use a very small font size for a simple html report
containing a dfSummary(). For this, we create a .css file (with the
name of our choosing) which contains the following class definition:
.tiny-text {
font-size: 8px;
}
Then we use print()’s custom.css argument to specify to location of
our newly created CSS file (results not shown):
print(dfSummary(tobacco), custom.css = 'path/to/custom.css',
table.classes = 'tiny-text', file = "tiny-tobacco-dfSummary.html")
10. Creating Shiny apps
To successfully include summarytools functions in Shiny apps,
- use html rendering
- set
bootstrap.css = FALSEto avoid interacting with the app’s layout - set
headings = FALSEin case problems arise - adjust graph sizes with
print()’sgraph.magnifparameter or with thedfSummary.graph.magnifglobal option - if
dfSummary()tables are too wide, omit a column or two (valid.colandvarnumbers, for instance) - if the results are still unsatisfactory, set column widths manually
with
print()’scol.widthsparameter
Example (results not shown)
print(dfSummary(somedata, varnumbers = FALSE, valid.col = FALSE,
graph.magnif = 0.8),
method = 'render',
headings = FALSE,
bootstrap.css = FALSE)
11. Graphs in Markdown dfSummaries
When using dfSummary() in an Rmd document using markdown styling
(as opposed to html rendering), three elements are needed in order to
display the png graphs properly:
1 - plain.ascii must be set to FALSE
2 - style must be set to “grid”
3 - tmp.img.dir must be defined
Why the third element? Although R makes it really easy to create temporary files and directories, they do have long pathnames, especially on Windows. Unfortunately, Pandoc determines the final (rendered) column widths by counting characters in a cell, even if those characters are paths pointing to images.
At this time, there seems to be only one solution around this problem: cut down on characters in image paths. So instead of this:
+-----------+---------------------------------------------------------------------+---------+
| Variable | Graph | Valid |
+===========+=====================================================================+=========+
| gender\ |  | 978\ |
| [factor] | | (97.8%) |
+----+---------------+------------------------------------------------------------+---------+
…we aim for this:
+---------------+----------------------+---------+
| Variable | Graph | Valid |
+===============+======================+=========+
| gender\ |  | 978\ |
| [factor] | | (97.8%) |
+---------------+----------------------+---------+
CRAN policies are really strict when it comes to writing content in the user directories, or anywhere outside R’s temporary zone (for good reasons). So the users need to set this location themselves, therefore consenting to having content written outside R’s predefined temporary zone.
On Mac OS and Linux, using “/tmp” makes a lot of sense: it’s a short path, and it’s self-cleaning. On Windows, there is no such convenient directory, so we need to pick one – be it absolute (“/tmp”) or relative (“img”, or simply “.”). Two things are to be kept in mind: it needs to be short (5 characters max) and it needs to be cleaned up manually.
12. Translations
Thanks to the R community’s efforts, the following languages can be used, in addition to English (default): French (fr), Portuguese (pt), Russian (ru), Spanish (es), and Turkish (tr).
To switch languages, simply use
st_options(lang = "fr")
All output from the core functions will now use that language:
freq(iris$Species)
## setting plain.ascii to FALSE
Tableau de fréquences
iris$Species
Type: Facteur
| Fréq. | % Valide | % Valide cum. | % Total | % Total cum. | |
|---|---|---|---|---|---|
| setosa | 50 | 33.33 | 33.33 | 33.33 | 33.33 |
| versicolor | 50 | 33.33 | 66.67 | 33.33 | 66.67 |
| virginica | 50 | 33.33 | 100.00 | 33.33 | 100.00 |
| <NA> | 0 | 0.00 | 100.00 | ||
| Total | 150 | 100.00 | 100.00 | 100.00 | 100.00 |
12.1 Non-UTF-8 Locales
On most Windows systems, it will be necessary to change the LC_CTYPE
element of the locale settings if the character set is not included in
the system’s default locale. For instance, in order to get good results
with the Russian language in a “latin1” environment, we need to do the
following:
Sys.setlocale("LC_CTYPE", "russian")
st_options(lang = 'ru')
Then to go back to default settings:
Sys.setlocale("LC_CTYPE", "")
st_options(lang = "en")
12.2 Defining and Using Custom Translations
Using the function use_custom_lang(), it is possible to add your own
set of translations. To achieve this, get the csv
template,
customize the +/- 70 items, and call use_custom_lang(), giving it as
sole argument the path to the edited csv template. Note that such
custom translations will not persist across R sessions. This means that
you should always have this csv file handy for future use.
12.3 Defining Specific Keywords
Sometimes, all you might want to do is change just a few keywords – for
instance, you could prefer using “N” instead of “Freq” in the title row
of freq() tables. For this, use define_keywords(). Calling this
function without any arguments will bring up, on systems that support
graphical devices (the vast majority, that is), an editable window
allowing to modify only the desired item(s).
After closing the edit window, you will be able to export the resulting
“custom language” into a csv file that you can reuse in the future by
calling use_custom_lang().
It is also possible to programmatically define one or several keywords
using define_keywords(). For instance:
define_keywords(freq = "N")
See ?define_keywords for more details.
13. Additional Software Installations
Required Software on Mac OS
Open a terminal window and enter the following:
brew install [email protected]
If you do not have brew installed, simply enter this command in the terminal:
/usr/bin/ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)"
If you’re using Mac OS X version 10.8 (Mountain Lion) or more recent versions, you’ll need to download the .dmg image from xquartz.org and add it to your Applications folder.
Back to installation instructions
Required Software for Debian / Ubuntu / Linux Mint
Magick++
sudo apt install libmagick++-dev
Back to installation instructions
Required Software for Older Ubuntu Versions
This applies only if you are using Ubuntu Trusty (14.04) or Xenial (16.04).
sudo add-apt-repository -y ppa:opencpu/imagemagick
sudo apt-get update
sudo apt-get install -y libmagick++-dev
Back to installation instructions
Required Software for Fedora / Red Had / CentOS
Magick++
sudo yum install ImageMagick-c++-devel
Back to installation instructions
Required Software for Solaris
pkgadd -d http://get.opencsw.org/now
/opt/csw/bin/pkgutil -U
/opt/csw/bin/pkgutil -y -i imagemagick
/usr/sbin/pkgchk -L CSWimagemagick
Back to installation instructions
14. Conclusion
The package comes with no guarantees. It is a work in progress and feedback is always welcome. Please open an issue on GitHub if you find a bug or wish to submit a feature request.
Stay Up to Date, and Get Involved!
For a preview of what’s coming in the next release, have a look at the development branch.
So far, I’ve worked a lot on my own on this project. Now I need your help to make it more of a collective effort. Check out the Wiki and don’t hesitate to post in the Discussions section.
15. Sponsors
A big thanks to people who made donations!
- Ashirwad Barnwal
- David Thomas
- Peter Nilsson
- Ross Dunne
- Igor Rubets
If you find summarytools useful and want to support its development, please consider making a small donation using the PayPal button.