compomics-utilities
compomics-utilities publication:
- Barsnes et al: BMC Bioinformatics. 2011 Mar 8;12(1):70.
- If you use compomics-utilities as part of a paper, please include the reference above.
What is compomics-utilities?
The Computational Omics and Systems Biology Group develops various bioinformatics tools for analyzing omics data.
compomics-utilities is a library containing code shared by many of our research projects, amongst others containing panels for visualizing spectra and chromatograms and objects for representing peptides and proteins etc. We believe that this library can be of use to other research groups doing computational proteomics, and have therefore made it available as open source.
Projects using compomics-utilities
Using compomics-utilities
For example code and demos showing how the compomics-utilities library can be used in your project see Examples. Also check the JavaDoc.
Download
We strongly recommend Maven when using compomics-utilities, as this makes sure that one gets all the dependencies right. See Maven Dependency below.
All compomics-utilities builds are available here.
See also the JavaDoc.
If you for some reason cannot use Maven and need a complete build of compomics-utilities, please let us know and we will send you the latest build as a zip file.
Maven Dependency
compomics-utilities is available for use in Maven projects:
<dependency>
<groupId>com.compomics</groupId>
<artifactId>utilities</artifactId>
<version>X.Y.Z</version>
</dependency>
<repositories>
<!-- Compomics Genesis Maven 2 repository -->
<repository>
<id>genesis-maven2-repository</id>
<name>Genesis maven2 repository</name>
<url>http://genesis.UGent.be/maven2</url>
<layout>default</layout>
</repository>
<!-- old EBI repository -->
<repository>
<id>ebi-repo</id>
<name>The EBI internal repository</name>
<url>http://www.ebi.ac.uk/~maven/m2repo</url>
</repository>
<!-- EBI repository -->
<repository>
<id>pst-release</id>
<name>EBI Nexus Repository</name>
<url>http://www.ebi.ac.uk/Tools/maven/repos/content/repositories/pst-release</url>
</repository>
</repositories>
Update the version number to latest released version, see the Maven repository.
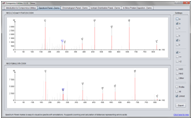
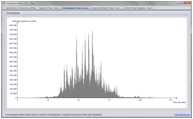
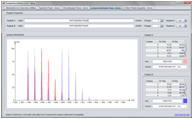
Screenshots
(Click on figure to see the full size version)
 |
 |
 |
 |
 |
