Shen-Lab / Deepaffinity
Programming Languages
Projects that are alternatives of or similar to Deepaffinity
DeepAffinity
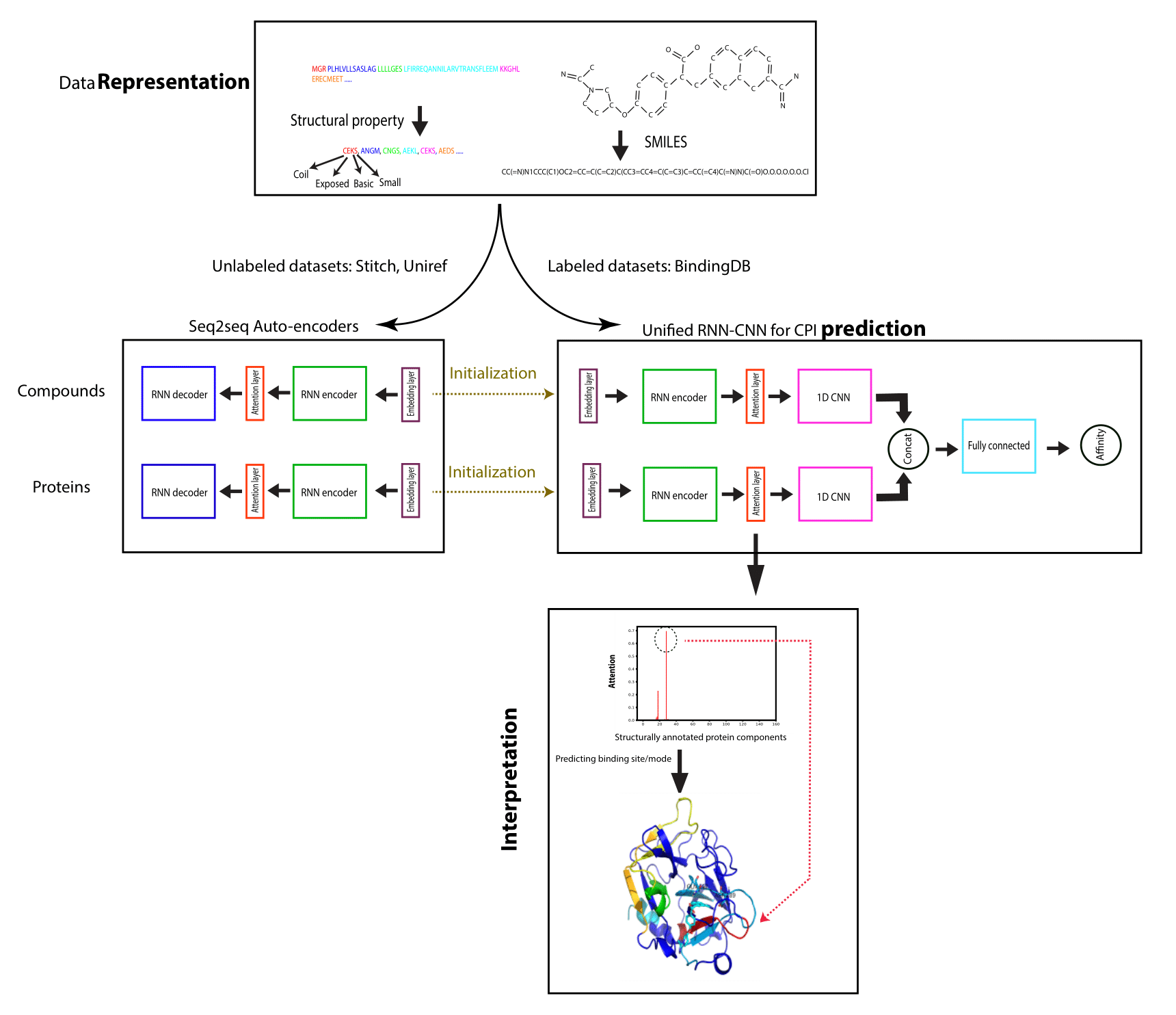
Drug discovery demands rapid quantification of compound-protein interaction (CPI). However, there is a lack of methods that can predict compound-protein affinity from sequences alone with high applicability, accuracy, and interpretability. We present a integration of domain knowledges and learning-based approaches. Under novel representations of structurally-annotated protein sequences, a semi-supervised deep learning model that unifies recurrent and convolutional neural networks has been proposed to exploit both unlabeled and labeled data, for jointly encoding molecular representations and predicting affinities. Performances for new protein classes with few labeled data are further improved by transfer learning. Furthermore, novel attention mechanisms are developed and embedded to our model to add to its interpretability. Lastly, alternative representations using protein sequences or compound graphs and a unified RNN/GCNN-CNN model using graph CNN (GCNN) are also explored to reveal algorithmic challenges ahead.
Pre-requisite:
- Tensorflow-gpu v1.1
- Python 3.6
- TFLearn v0.3
- Scikit-learn v0.19
- Anaconda 3/5.0.0.1
- We have already provided our environment list as environment.yml. You can create your own environment by:
conda env create -n envname -f environment.yml
Table of contents:
- data_script: Contain the supervised learning datasets(pIC50, pKi, pEC50, and pKd)
- Seq2seq_models: Contain auto-encoder seq2seq models and their data for both SPS and SMILE representations
- baseline_models: Contain shallow models for both Pfam/pubchem features and features generated from the encoder part of seq2seq model
- Separate_models: Contain deep learning model for features generated from the encoder part of seq2seq model
-
Joint_models: Contain all the joint models including:
- Separate attention mechanism
- Marginalized attention mechanism
- Joint attention mechanism
- Graph convolution neural network (GCNN) with separate attention mechanism
Testing the model
To test DeepAffinity for new dataset, please follow the steps below:
- Download the checkpoints trained based on training set of IC50 from the following link
- Download the checkpoints trained based on the whole dataset of IC50 from the following link
- Download the checkpoints trained based on the whole dataset of Kd from the following link
- Download the checkpoints trained based on the whole dataset of Ki from the following link
- Put your data in folder "Joint_models/joint_attention/testing/data"
- cd Joint_models/joint_attention/testing/
- Run the Python code joint-Model-test.py
You may use the script to run our model in one command. The details can be found in our manual (last updated: Apr. 9, 2020).
(Aug. 21, 2020) We are now providing SPS (Structure Property-annotated Sequence) for all human proteins! zip (Credit: Dr. Tomas Babak at Queens University). Columns: 1. Gene identifier 2. Protein FASTA 3. SS (Scratch) 4. SS8 (Scratch) 5. acc (Scratch) 6. acc20 7. SPS
P.S. Considering the distribution of protein sequence lengths in our training data, our trained checkpoints are recommended for proteins of lengths between tens and 1500.
Re-training the seq2seq models for new dataset:
(Added on Jan. 18, 2021) To re-train the seq2seq models for new dataset, please follow the steps below:
- Use the translate.py function in any of the seq2seq models with the following important arguments:
- data_dir: data directory where includes all the data
- train_dir: training directory where all the checkpoints will be saved in.
- from_train_data: source training data which will be translated from.
- to_train_data: target training data which will be translated to (can be the same with from_train_data if doing auto-encoding which we used in the paper).
- from_dev_data: source validation data which will be translated from.
- to_dev_data: target validation data which will be translated to (can be the same with from_dec_data if doing auto-encoding which we used in the paper).
- num_layers: Number of RNN layers (default 2)
- batch_size: Batch size (default 256)
- num_train_step: number of training steps (default 100K)
- size: the size of hidden dimension for RNN models (default 256)
- SPS_max_length (SMILE_max_length): maximum length of SPS (SMILE)
- Suggestion for using seq2seq models:
- For protein encoding: seq2seq_part_FASTA_attention_fw_bw
- For compound encoding: seq2seq_part_SMILE_attention_fw_bw
- Example of running for proteins: python translate.py --data_dir ./data --train_dir ./checkpoints --from_train_data ./data/FASTA_from.txt --to_train_data ./data/FASTA_to.txt --from_dev_data ./data/FASTA_from_dev.txt --to_dev_data ./data/FASTA_to_dev.txt --SPS_max_length 152
- Once the training is done, you should copy the parameters' weights cell_*.txt, embedding_W.txt, *_layer_states.txt in the joint_attention/joint_fixed_RNN/data/prot_init which will be used for the next step, supervised training in the joint attention model (you can do the same for separate and marginalized attention models as well)
Note:
We recommend referring to PubChem for canonical SMILES for compounds.
Citation:
If you find the code useful for your research, please consider citing our paper:
@article{karimi2019deepaffinity,
title={DeepAffinity: interpretable deep learning of compound--protein affinity through unified recurrent and convolutional neural networks},
author={Karimi, Mostafa and Wu, Di and Wang, Zhangyang and Shen, Yang},
journal={Bioinformatics},
volume={35},
number={18},
pages={3329--3338},
year={2019},
publisher={Oxford University Press}
}
Contacts:
Yang Shen: [email protected]
Di Wu: [email protected]
Mostafa Karimi: [email protected]