MetaOmGraph
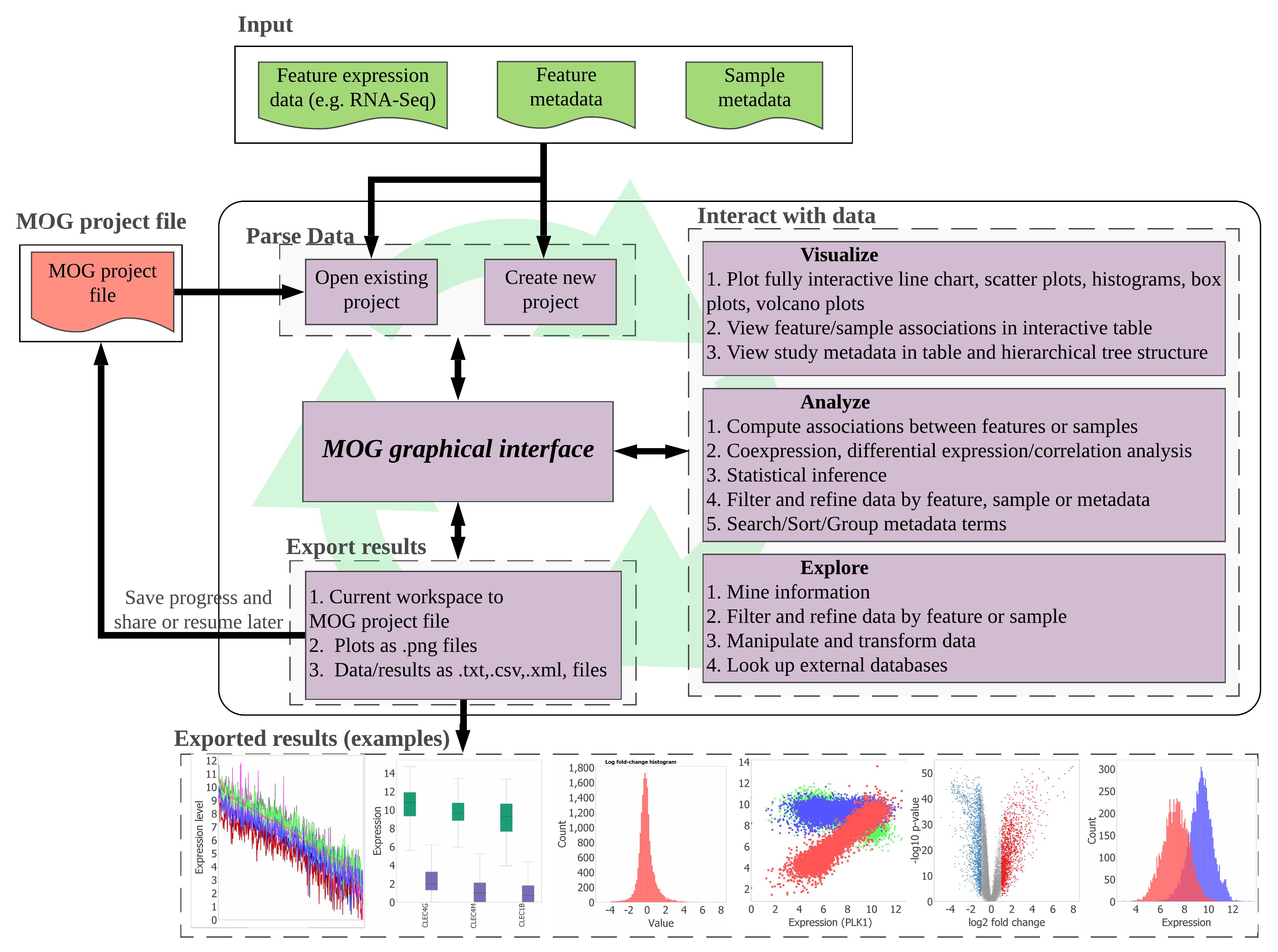
MetaOmGraph (MOG) is a Java software to interactively explore and visualize large datasets. MOG overcomes the challenges posed by big size and complexity of big datasets by efficient handling of data files by using a combination of data indexing and buffering schemes. By incorporating metadata, MOG adds another dimension to the analyses and provides flexibility in data exploration. MOG allows users to explore their own data on their local machines. The user can save the progress at any stage of analysis to a MOG project file. Saved MOG projects can be shared, reused, and included in publications. MOG is user-centered software, designed for all types of data, but specialized for expression data. It combines the ability to analyze very large data sets in real time with metadata analysis, statistical analysis, list-making, and graphing capabilities.
Citation
Examples
Line Chart
Box Plot
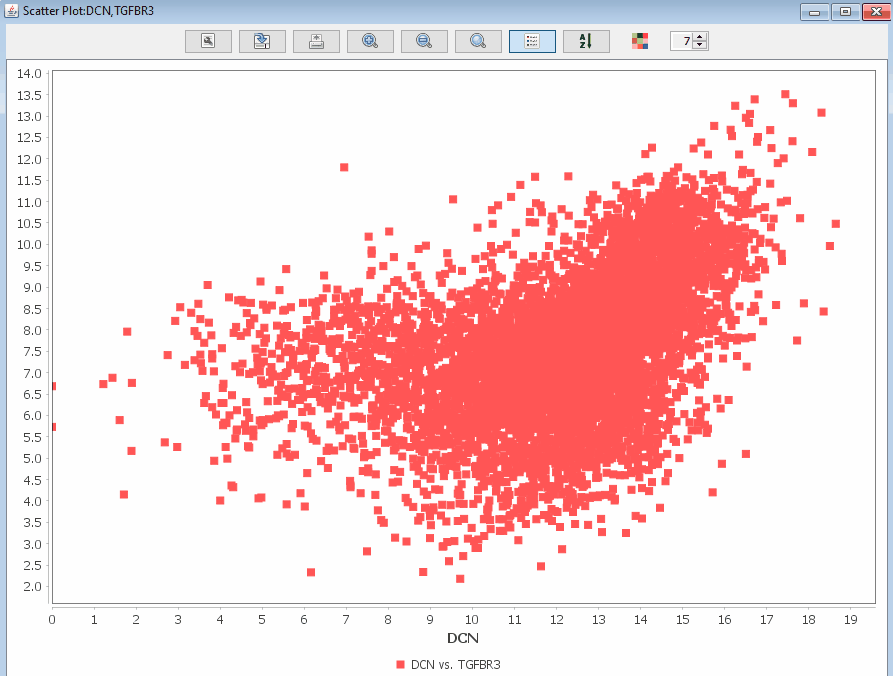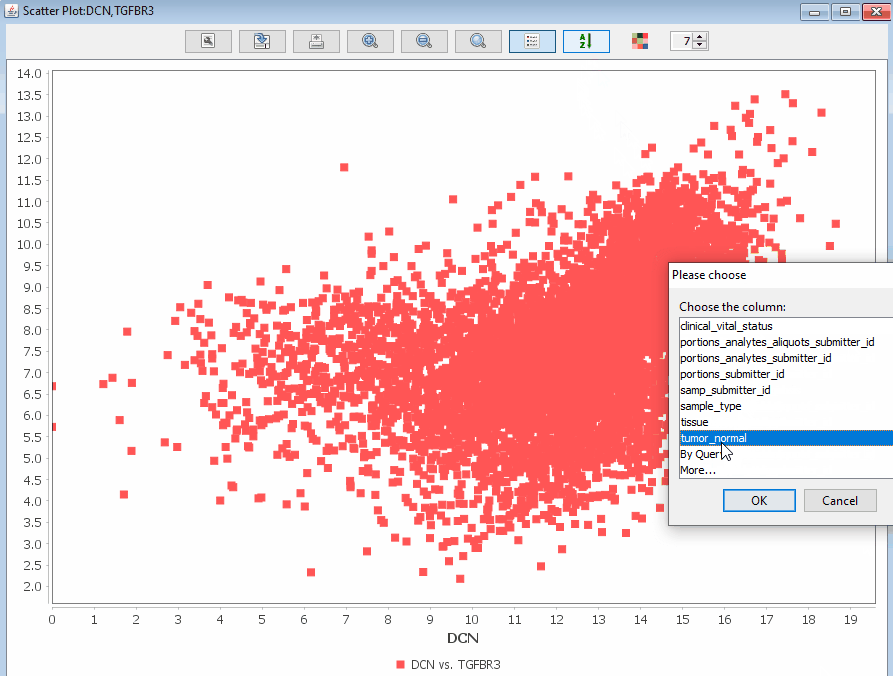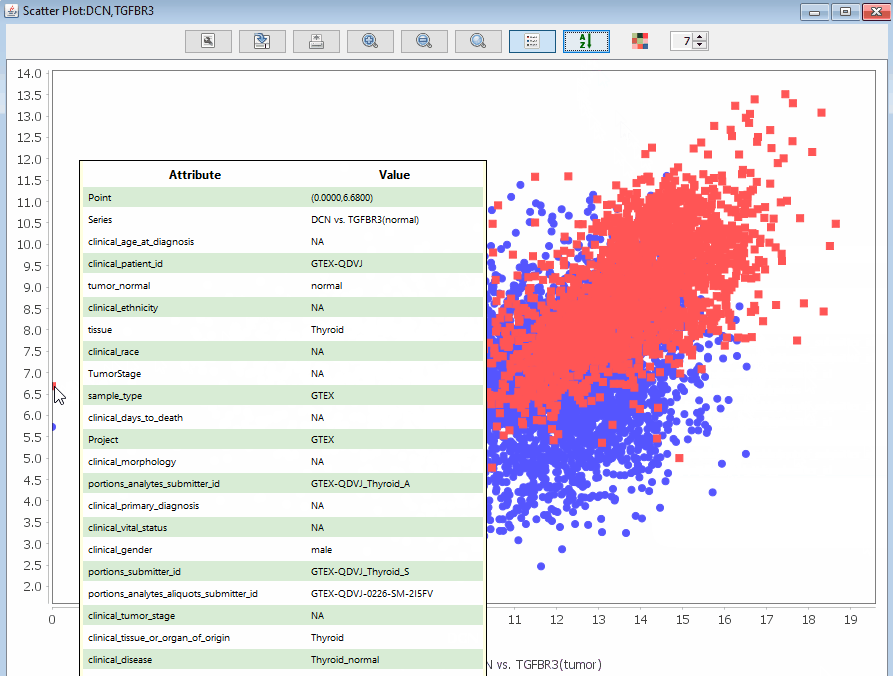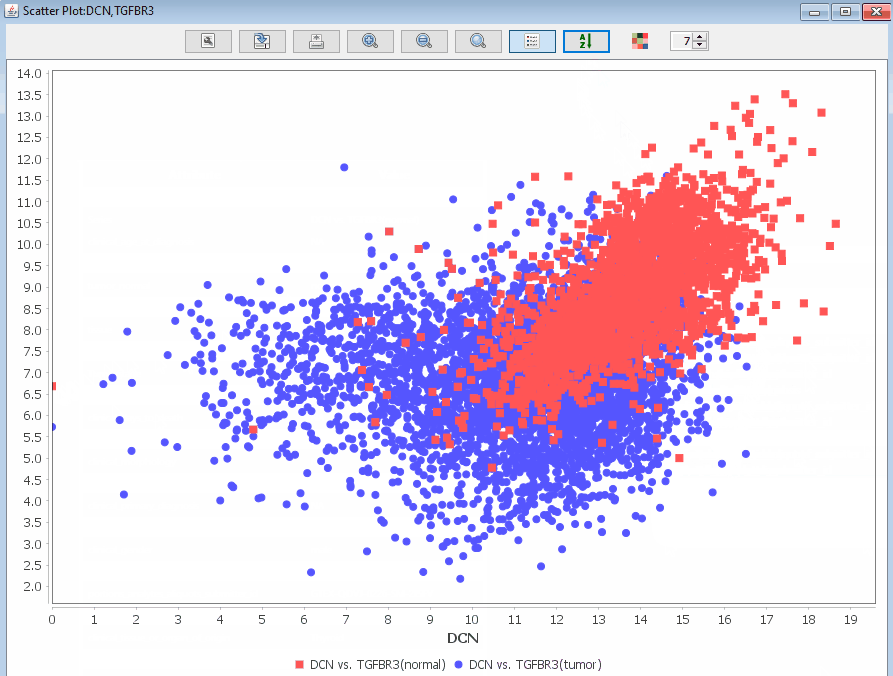
Scatter Plot
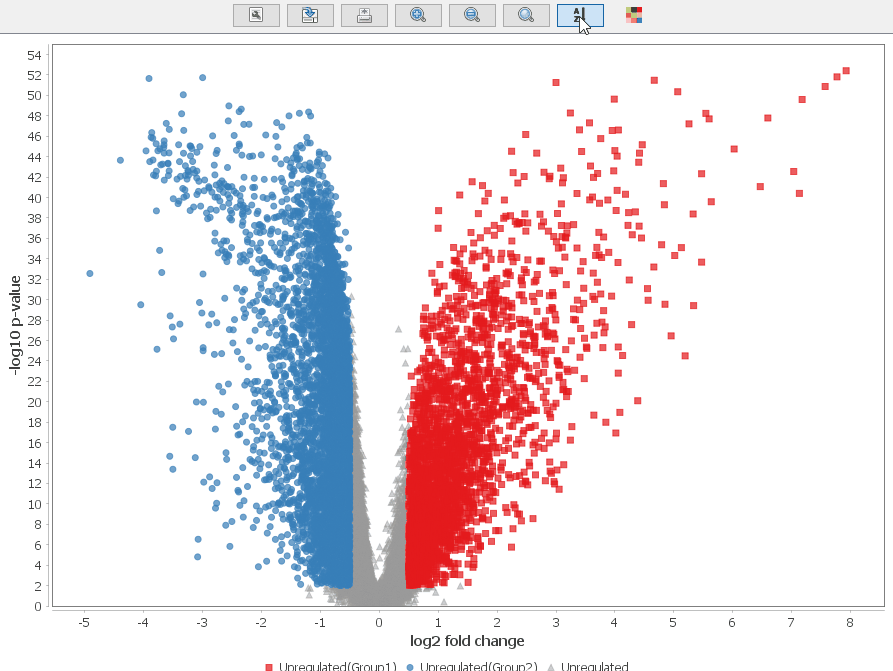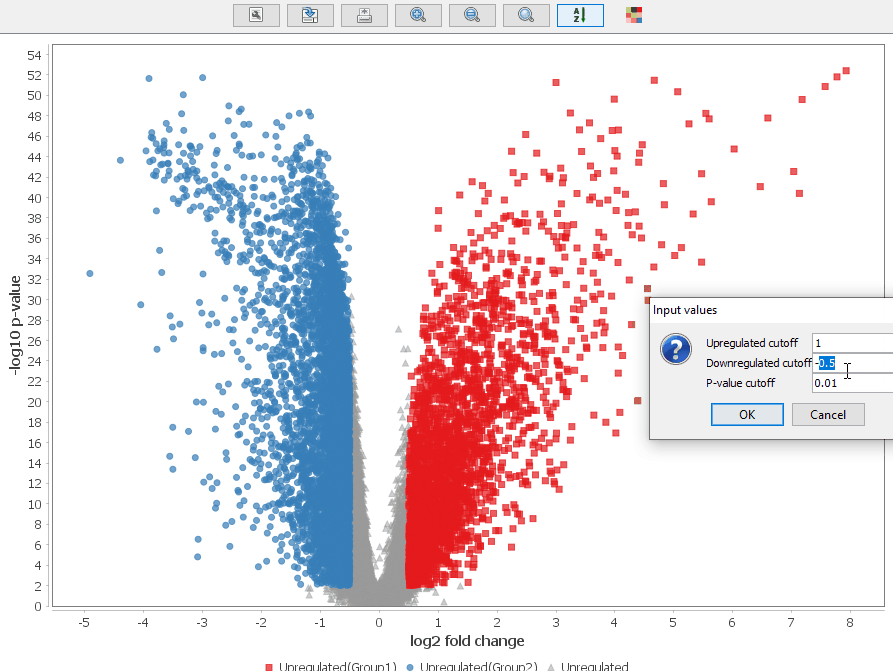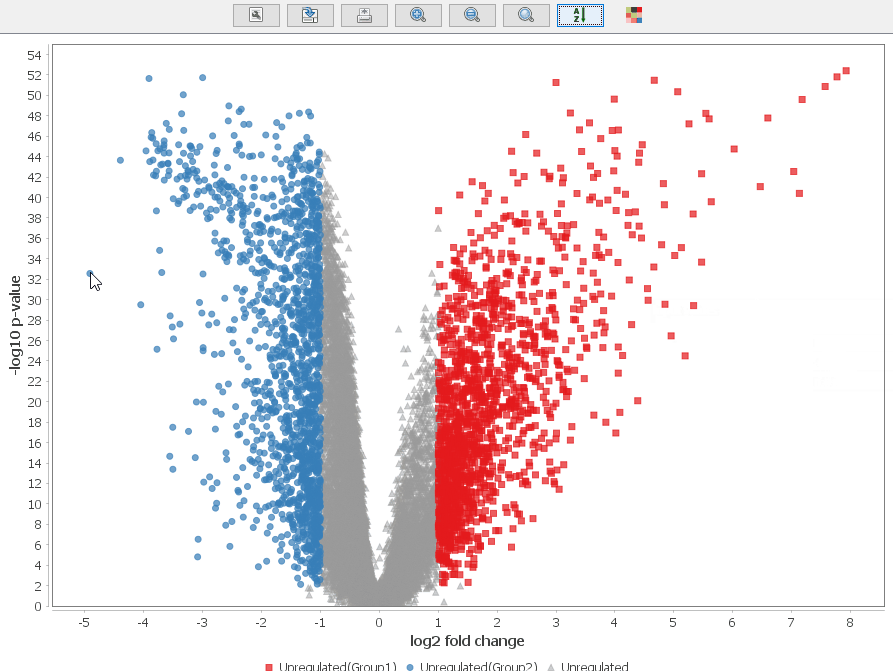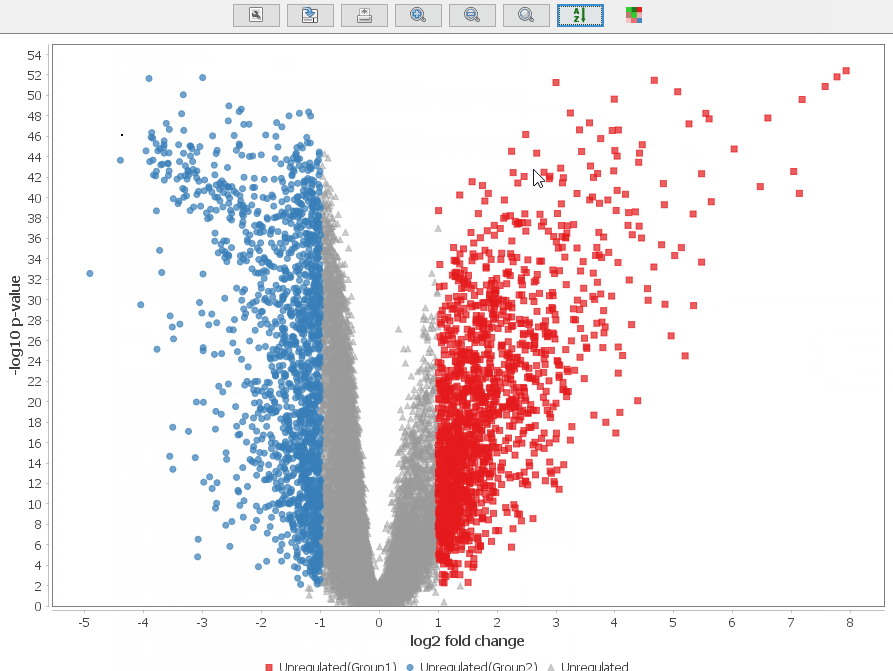
Volcano Plot
Histogram
Filtering Samples
Getting Started
Download executable from: http://metnetweb.gdcb.iastate.edu/MetNet_MetaOmGraph.htm
User guide is available at: https://github.com/urmi-21/MetaOmGraph/blob/master/manual/MOG_User_Guide.pdf
Prerequisites
- Java Runtime Environment 8 (or higher)
- R 3.4 (or higher) [optional]
Installing
MOG is freely available to download from http://metnetweb.gdcb.iastate.edu/MetNet_MetaOmGraph.htm. Click the download button, and then download the .zip file. Unzip the downloaded file to get a .jar file, this is the MOG program.
DOUBLE CLICK on the .jar file icon to start MOG.
Contributing
Please see CONTRIBUTING.md
Contributors
Current developers
- Urminder Singh
- Sumanth Kaliki
- Kumara Sri Harsha
Initial developers
- Manhoi Hur
- Nick Ransom
Getting Help
If you encounter an error/bug, please report a minimal reproducible example on github. For questions and other discussion, please get in touch with the developers via github or email.
License
This project is licensed under the MIT License - see the LICENSE file for details
Some Java libraries MOG uses
References
If you use MetaOmGraph in your research, please cite the paper Urminder Singh, Manhoi Hur, Karin Dorman, Eve Syrkin Wurtele, MetaOmGraph: a workbench for interactive exploratory data analysis of large expression datasets, Nucleic Acids Research.
If you use Mutual Information module in MetaOmGraph, please also cite Daub et al. Estimating mutual information using B-spline functions--an improved similarity measure for analysing gene expression data