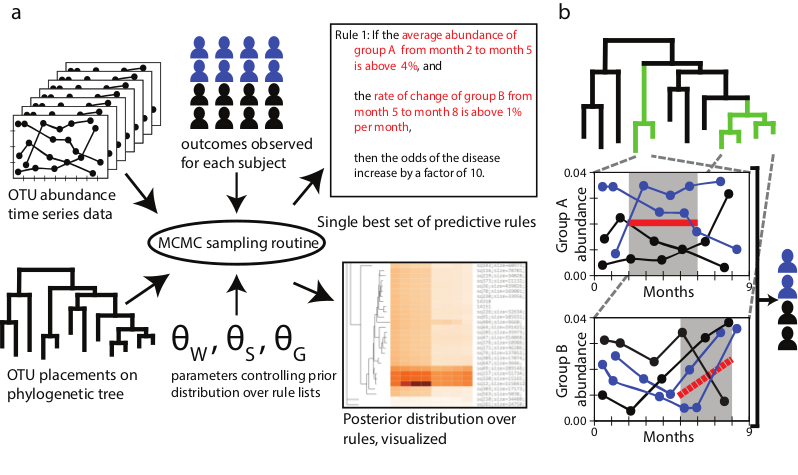
The Microbiome Interpretable Temporal Rule Engine
MITRE learns predictive models of patient outcomes from microbiome time-series data in the form of short lists of interpretable rules.
See an example of MITRE's interactive visualization output.
Installation
Python 2.7 and a C compiler are required to install MITRE.
From PyPI (recommended):
$ pip install mitre
From source:
$ git clone https://github.com/gerberlab/mitre.git
$ pip install mitre/
To check that installation was successful, run
$ mitre --test
A series of status messages should be displayed, followed by 'Test problem completed successfully.'
If you don't have the 'pip' command
Recent versions of Python 2.7 provide pip by default, but the version of Python installed by default on OSX systems, for example, is an exception. Running
$ sudo easy_install pip
should fix this if you are an administrator, but a better solution, which does not require administrator access, is to install your own Python interpreter. We recommend the Anaconda distribution which installs key scientific python libraries by default and provides an improved package management and installation system.
Supported platforms
Only Mac and Linux systems are supported at this time.
Quick start
MITRE operation is controlled by a configuration file. To try it out, copy the following into a file called 'demo.cfg':
[general]
verbose = True
[data]
load_example = bokulich
[preprocessing]
min_overall_abundance = 10
min_sample_reads = 5000
trim_start = 0
trim_stop = 375
density_filter_n_samples = 1
density_filter_n_intervals = 12
density_filter_n_consecutive = 2
take_relative_abundance = True
aggregate_on_phylogeny = True
temporal_abundance_threshold = 0.0001
temporal_abundance_consecutive_samples = 3
temporal_abundance_n_subjects = 10
discard_surplus_internal_nodes = True
[model]
n_intervals = 12
t_min = 1.0
t_max = 180.0
[sampling]
total_samples = 300
[postprocessing]
burnin_fraction = 0.05
bayes_factor_samples = 1000
quick_summary = True
full_summary = True
gui_output = True(If you've downloaded the MITRE source code, you can copy demo.cfg from the root directory.)
Then run
$ mitre demo.cfg
in the same directory. It should take 15-20 minutes to run. Here's what will happen:
MITRE will load data from Bokulich, N. A., et al., "Antibiotics, birth mode, and diet shape microbiome maturation during early life." Science Translational Medicine 8(343): 343ra82, which is packaged with MITRE.
Then, it will apply a series of filters:
- excluding OTUs and samples with too few associated reads
- truncating the experiment at day of life 375,
- Dividing the 375-day study into 12 intervals and discarding subjects without at least 1 sample in any consecutive 2 intervals.
Next, counts data will be converted to relative abundance, and new variables representing the aggregated abundance of every subtree in a phylogenetic tree relating the OTUs will be created. Variables not exceeding an abundance threshold will be dropped.
A MITRE model object will be set up and 300 samples will be drawn from the posterior distribution over the space of valid rule sets relating the microbiome time series data to the outcome of interest.
Three output files summarizing the samples will be written. In this demo, we aren't drawing enough samples to get reliable statistics, so the results may vary. But bokulich_diet_quick_summary.txt might look something like this:
POINT SUMMARY:
Rule list with 1 rules (overall likelihood -7.14):
Rule 0 (coefficient 11.5 (5.28 -- 13.2)):
Odds of positive outcome INCREASE by factor of 9.57e+04 (196 - 5.61e+05), if:
Between time 93.750 and time 156.250, variable 13231 average is above 0.1309
This rule applies to 9/35 (0.257) subjects in dataset, 9/9 with positive outcomes (1.000).
Constant term (coefficient -2.81 (-4.1 -- -1.72)):
Positive outcome probability 0.0566 (0.0163 -- 0.152) if no other rules apply
This is the single best estimated rule set. It indicates that subjects with a high abundance of group 13231 in the indicated time window are likelier to have been fed predominantly a formula-based diet. In bokulich_diet_variable_annotations.txt, we can look up group 13231 and learn it's "a clade within phylum Firmicutes,including representatives of class Clostridia, Bacilli"- which isn't too enlightening on its own, but the same line lists the leaves of the tree that belong to this group; looking them up in turn, we find this group includes mostly OTUs from the genera Clostridia and Blautia. Ranges in parentheses are 95% confidence intervals.
Looking farther down the file, we find a confusion matrix showing that this rule set correctly identifies 9 of the 11 subjects in the group with a formula-dominant diet, with no false positives.
For an interactive representation, open bokulich_diet_visualization.html and click on the heat map to explore high-probability detectors. It might look like a noiser version of the example linked above.
For more details, see the user's manual and the text and supplementary note of the MITRE manuscript (reference below.)
References
Bogart, E., Creswell, R. & Gerber, G.K. "MITRE: inferring features from microbiota time-series data linked to host status." Genome Biol 20, 186 (2019).
Also available in earlier preprint form, with a title that communicates rather than obfuscates: "MITRE: predicting host status from microbiota time-series data", Elijah Bogart, Richard Creswell, and Georg K. Gerber.
License information
Copyright 2017-2019 Eli Bogart, Richard Creswell, the Gerber Lab, and Brigham and Women's Hospital. Released under the GNU General Public License version 3.0, see LICENSE.txt.