RivuletStudio / Rivuletpy
Programming Languages
Projects that are alternatives of or similar to Rivuletpy
Rivuletpy
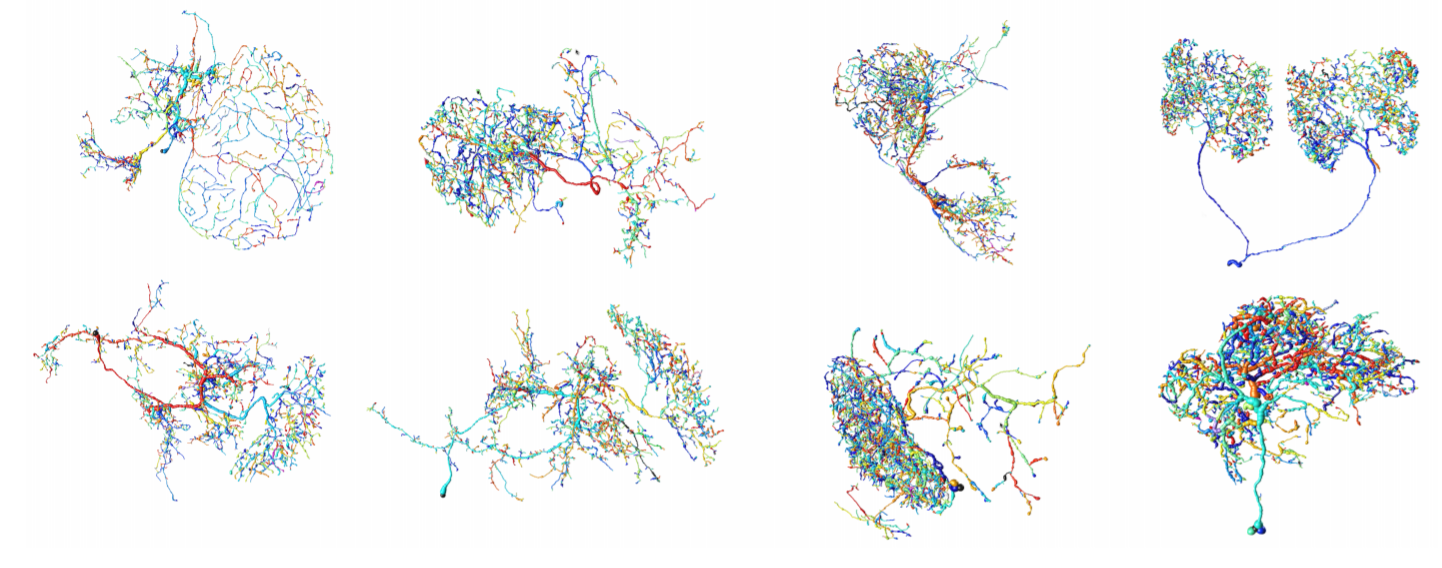
Example Neuron Tracings
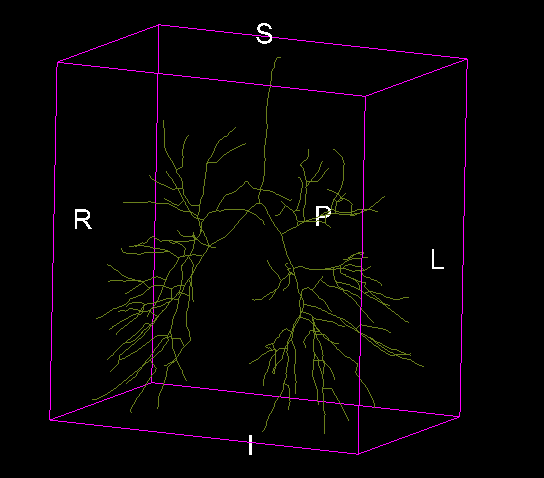
Example Lung Airway Tracing
Rivuletpy == Rivulet2
Rivuletpy is a Python3 toolkit for automatically reconstructing single neuron models from 3D microscopic image stacks & other tree structures from 3D medical images.
It is actively maintained and being used in industry scale image analysis applications.
The project was initiated in the BigNeuron project
The rtrace command is powered by the Rivulet2 algorithm published in IEEE Trans. TMI:
[1] S. Liu, D. Zhang, Y. Song, H. Peng and W. Cai, "Automated 3D Neuron Tracing with Precise Branch Erasing and Confidence Controlled Back-Tracking," in IEEE Transactions on Medical Imaging. URL: http://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=8354803&isnumber=4359023
PDF [https://www.biorxiv.org/content/biorxiv/early/2017/11/27/109892.full.pdf]
The predecessor Rivulet1 was published on Neuroinformatics:
[2] Siqi Liu, Donghao Zhang, Sidong Liu, Dagan Feng, Hanchuan Peng, Weidong Cai, "Rivulet: 3D Neuron Morphology Tracing with Iterative Back-Tracking", Neuroinformatics, Vol.14, Issue 4, pp387-401, 2016.
A C++ implementation of the Rivulet2 algorithm is also available in the lastest Vaa3D sources under the Rivulet Plugin (Not yet available in the released build). However you can build Vaa3D easily on Mac/Linux following the Vaa3D wiki carefully.
Issues / questions / pull requests
Issues should be reported to the Rivuletpy github repository issue tracker. The ability and speed with which issues can be resolved depends on how complete and succinct the report is. For this reason, it is recommended that reports be accompanied with a minimal but self-contained code sample that reproduces the issue, the observed and expected output, and if possible, the commit ID of the version used. If reporting a regression, the commit ID of the change that introduced the problem is also extremely valuable information.
Questions are also welcomed in the Rivuletpy github repository issue tracker.
If you put on a question label. We consider every question as an issue since it means we should have made things clearer/easier for the users.
Pull requests are definitely welcomed! Before you make a pull requests, please kindly create an issue first to discuss the optimal solution.
Installation
0. Setup the Anaconda environment
$ conda create -n riv python=python3.6 anaconda # We tested on 3.5 and 3.6. Other python versions >= 3.4 should also work
$ source activate riv
1. Setup the dependencies
To install rivuletpy with pip, you need to install the following packages manually beforehand since some dependencies of rivuletpy uses them in their setup scripts
numpy>=1.8.0scipy>=0.17.0Cython>=0.25.1tqdm-devlibtiff-devSimpleITK
(riv)$ conda install numpy scipy matplotlib cython tqdm
(riv)$ conda install -c conda-forge pylibtiff # Install pylibtiff for loading 3D tiff images
(riv)$ conda install -c simpleitk simpleitk # Install SimpleITK for the support of load mhd
2. Install Up-to-date Rivuletpy from source
Optionally you can install Rivuletpy from the source files
(riv)$ git clone https://github.com/RivuletStudio/rivuletpy.git
(riv)$ cd rivuletpy
(riv)$ python setup.py build
(riv)$ pip3 install .
Test Installation
In ./rivuletpy/
sh quicktest.sh
This will download a simple neuron image and perform a neuron tracing with rivulet2 algorithm. If you encountered any issues while installing Rivuletpy, you are welcome to raise an issue for the developers in the issue tracker
Usage
- Reconstruct single neuron file.
The script rtrace command will be installed
$ rtrace --help
usage: rtrace [-h] -f FILE [-o OUT] [-t THRESHOLD] [-z ZOOM_FACTOR]
[--save-soma] [--no-save-soma] [--speed]
[--quality] [--no-quality] [--clean] [--no-clean] [--silent]
[--no-silent] [-v] [--no-view]
[--tracing_resolution TRACING_RESOLUTION] [--vtk]
Arguments to perform the Rivulet2 tracing algorithm.
optional arguments:
-h, --help show this help message and exit
-f FILE, --file FILE The input file. A image file (*.tif, *.nii, *.mat).
-o OUT, --out OUT The name of the output file
-t THRESHOLD, --threshold THRESHOLD
threshold to distinguish the foreground and
background. Default 0. If threshold<0, otsu will be
used.
-z ZOOM_FACTOR, --zoom_factor ZOOM_FACTOR
The factor to zoom the image to speed up the whole
thing. Default 1.
--save-soma Save the automatically reconstructed soma volume along
with the SWC.
--no-save-soma Don't save the automatically reconstructed soma volume
along with the SWC (default)
--speed Use the input directly as speed image
--quality Reconstruct the neuron with higher quality and
slightly more computing time
--no-quality Reconstruct the neuron with lower quality and slightly
more computing time
--clean Remove the unconnected segments (default). It is
relatively safe to do with the Rivulet2 algorithm
--no-clean Keep the unconnected segments
--silent Omit the terminal outputs
--no-silent Show the terminal outputs & the nice logo (default)
-v, --view View the reconstructed neuron when rtrace finishes
--no-view
--tracing_resolution TRACING_RESOLUTION
Only valid for mhd input files. Will resample the mhd
array into isotropic resolution before tracing.
Default 1mm
--vtk Store the world coordinate vtk format along with the
swc
Example Usecases with single neurons in a TIFF image
$ rtrace -f example.tif -t 10 # Simple like this. Reconstruct a neuron in example.tif with a background threshold of 10
$ rtrace -f example.tif -t 10 --quality # Better results with longer running time
$ rtrace -f example.tif -t 10 --quality -v # Open a 3D swc viewer after reconstruction
Example Usecases with general tree structures in a mhd image
$ rtrace -f example.mhd -t 10 --tracing_resolution 1.5 --vtk # Perform the tracing under an isotropic resolution of 1.5mmx1.5mmx1.5mm and output a vtk output file under the world coordinates along side the swc.
$ rtrace -f example.mhd -t 10 --tracing_resolution 1.5 --vtk --speed # Use the input image directly as the source of making speed image. Recommended if the input mhd is a probablity map of centerlines.
Please note that Rivulet2 is powerful of handling the noises, a relatively low intensity threshold is preferred to include all the candidate voxels.
- Compare a swc reconstruction against the manual ground truth
$ compareswc --help
usage: compareswc [-h] --target TARGET --groundtruth GROUNDTRUTH
[--sigma SIGMA]
Arguments for comparing two swc files.
optional arguments:
-h, --help show this help message and exit
--target TARGET The input target swc file.
--groundtruth GROUNDTRUTH
The input ground truth swc file.
--sigma SIGMA The sigma value to use for the Gaussian function in
NetMets.
$ compareswc --target r2_tracing.swc --groundtruth hand_tracing.swc
0.9970 0.8946 0.9865 1 3
The compareswc command outputs five numbers which are in order:
precision, recall, f1-score, No. connection error type A, No. connection error type B
FAQ
What if I see on Mac OS ImportError: Failed to find TIFF library. Make sure that libtiff is installed and its location is listed in PATH|LD_LIBRARY_PATH|..
Try
brew install libtiff
What if I see ...version `GLIBCXX_3.4.21' not found... when I run rtrace under Anaconda?
Try
(riv)$ conda install libgcc # Upgrades the gcc in your conda environment to the newest
What if I see Intel MKL FATAL ERROR: Cannot load libmkl_avx2.so or libmkl_def.so.?
Try to get rid of the mkl in your conda, it has been reported to cause many issues
(riv)$ conda install nomkl numpy scipy scikit-learn numexpr
(riv)$ conda remove mkl mkl-service
Dependencies
The build-time and runtime dependencies of Rivuletpy are: