Wentong-DST / Im2p
Programming Languages
Projects that are alternatives of or similar to Im2p
im2p
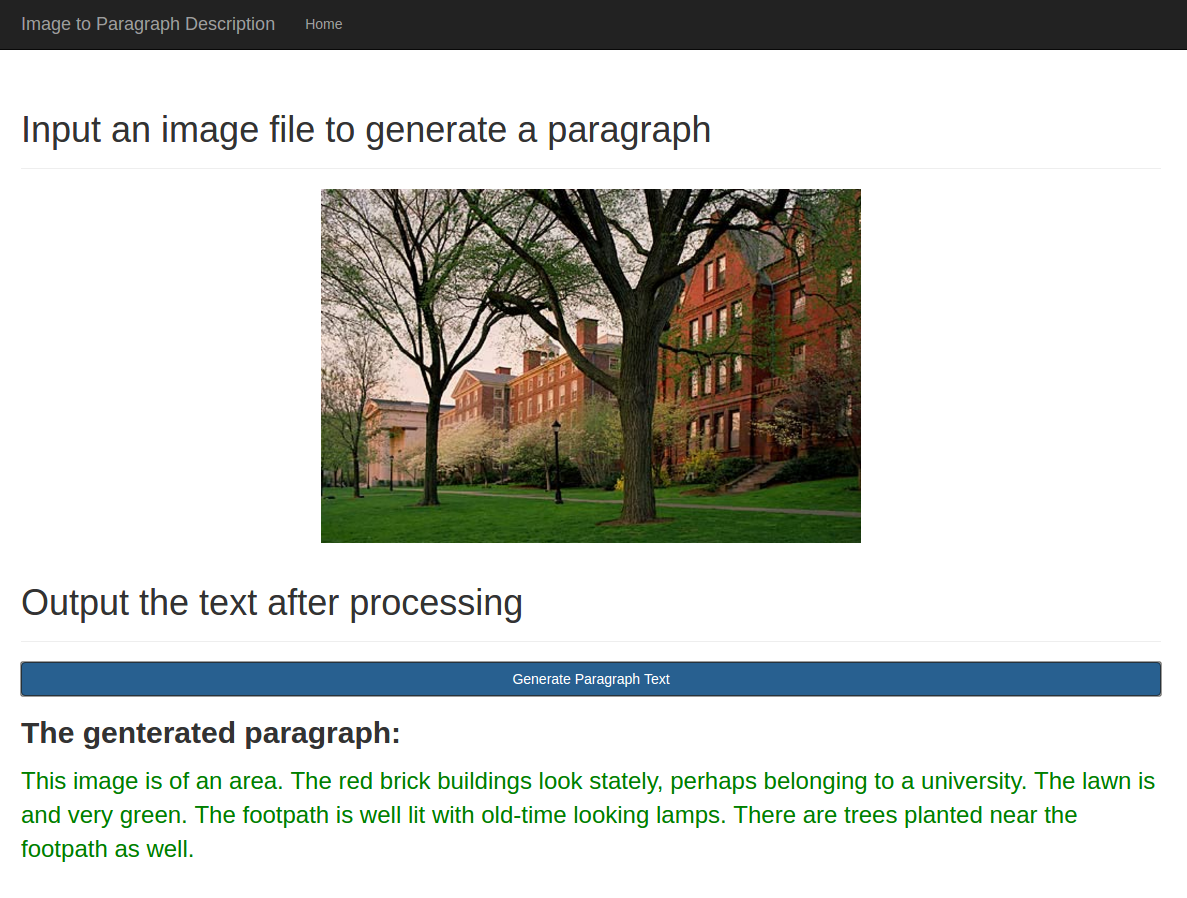
Tensorflow implement of paper: A Hierarchical Approach for Generating Descriptive Image Paragraphs.
Thanks to the original repo author chenxinpeng.
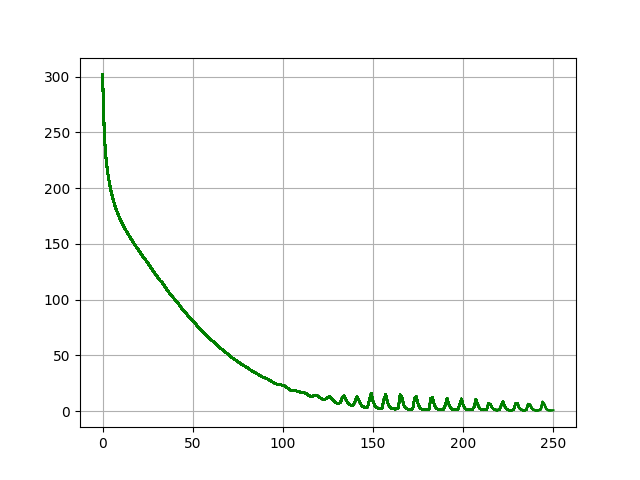
I haven't fine-tunning the parameters, but I achieve the metric scores (by chenxinpeng):
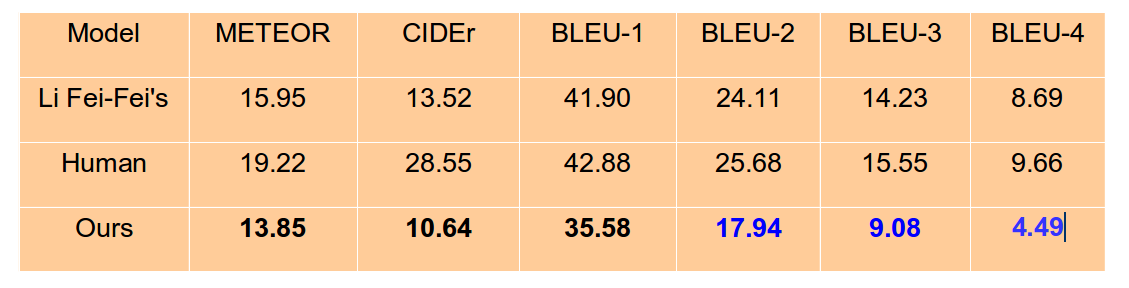
Please feel free to ask questions in Issues.
Step 1
Configure the torch running environment. Upgrade to Tensorflow v1.2 or above. Install Torch, recommend to use the approach described in Installing Torch without root privileges. Then deploy the running environment follow by densecap step by step.
To verify the running environment, run the script:
$ th check_lua_packages.lua
Also clone pycocoevalcap in same directory, but I have written some patches to fix some bugs, some replace [bleu.py, cider.py, meteor.py, rouge.py] with their corresponding files in pycocoevalcap folder.
Step 2
Download the VisualGenome dataset, we get the two files: VG_100K, VG_100K_2. According to the paper, we download the training, val and test splits json files. These three json files save the image names of train, validation, test data. We save them into data folder.
Running the script:
$ python split_dataset.py
We will get images from [VisualGenome dataset] which the authors used in the paper.
Step 3
Run the scripts:
$ python get_imgs_path.py
We will get three txt files: imgs_train_path.txt, imgs_val_path.txt, imgs_test_path.txt. They save the train, val, test images path.
After this, we use dense caption to extract features.
Step 4
Run the script:
$ ./download_pretrained_model.sh
We should download the pre-trained model: densecap-pretrained-vgg16.t7.
Then, according to the paper, we extract 50 boxes and the features from each image. So run the script:
$ ./extract_features.sh
in which the following command will be executed:
$ th extract_features.lua -boxes_per_image 50 -max_images -1 -input_txt imgs_train_path.txt \
-output_h5 ./data/im2p_train_output.h5 -gpu -1 -use_cudnn 0
Note that -gpu -1 means we are only using CPU when cudnn fails to run properly in torch.
Also note that my hdf5 module always crashes in torch, so I have to rewrite the features saving part in extract_features.lua by saving them directly to hard disk first, and then use h5py in Python to convert these features into hdf5 format. Run this script:
$ ./convert-to-hdf5.sh
Step 5
Run the script:
$ python parse_json.py
In this step, we process the paragraphs_v1.json file for training and testing, which looks like this:

We get the img2paragraph file in the ./data directory. Its structure is like this:

Step 6
Finally, we can train and test model, in the terminal:
$ CUDA_VISIBLE_DEVICES=0 ipython
>>> import HRNN_paragraph_batch.py
>>> HRNN_paragraph_batch.train()
After training, we can test the model:
>>> HRNN_paragraph_batch.test()
And then compute all evaluation metrics:
>>> HRNN_paragraph_batch.eval()