nmpowell / Mousemorph
Programming Languages
Projects that are alternatives of or similar to Mousemorph
MouseMorph
We're not quite done here, but please get in touch if you are interested: [email protected].
Tools for MRI mouse brain morphometry.
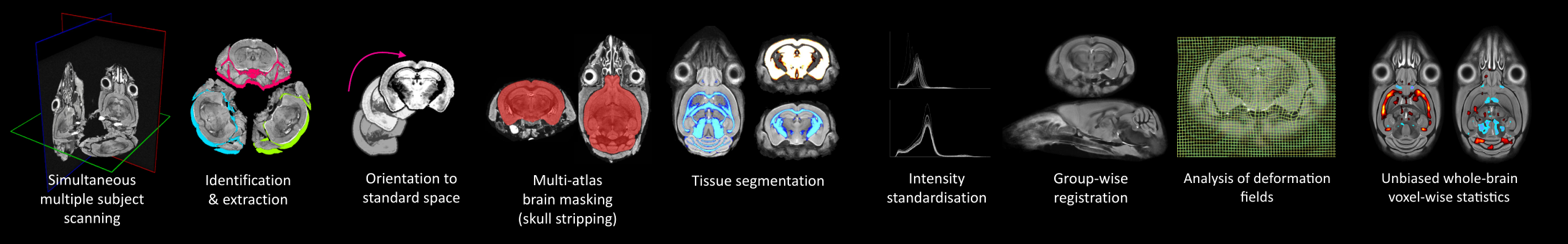
MouseMorph is a modular set of tools for automatically analysing mouse brain MRI scans. It enables fully automatic Voxel- and Tensor-Based Morphometry (VBM, TBM) on large cohorts of high-resolution images. It is employed at the UCL Centre for Advanced Biomedical Imaging (CABI) for phenotyping mice based on in vivo and ex vivo MRI. It has been tested for robustness on hundreds of mouse brain scans.
The primary distinction from clinically-focussed tools like SPM and FSL is a robust set of pre-processing steps, unique to -- or with customisations for -- the preclinical paradigm (mice and rats):
- Identification and extraction of multiple subjects from a single scan image
- Orientation to a standard space, from any initial orientation
- Mouse brain extraction (skull stripping / brain masking)
- Tissue segmentation, using accurate, mouse-specific priors
- Group-wise registration
Most of these steps are atlas-based (requiring prior knowledge). A few mouse atlases, fulfilling this requirement, are freely available to download (see links below). We aim to release more. For a further introduction, see the poster. For links to open and free wild-type mouse brain MRI data, see below.
Developed at the [email protected]) and others.
Phenotyping
If you are interested in using MouseMorph to assist a phenotyping study, please get in touch!
Multiple mouse brain MRI
- See the brain holder in this repository (stl folder), or at Figshare (please cite: https://dx.doi.org/10.6084/m9.figshare.1290771)
Setup
- Download, and install, NiftyReg, and NiftySeg. Ensure the executables are on your system path (Linux; Windows)
- You need at least Python 2.7. Try Anaconda, which includes the required NumPy, SciPy, Pandas, Matplotlib.
- You also need NiBabel. From a command line, run:
pip install nibabel - Download the MouseMorph repository as a .zip, and extract
- From the mousemorph directory, run:
python setup.py install
Publications
MouseMorph has been used in several peer-reviewed publications:
- Powell, NM., Modat, M., Cardoso, MJ., Ma, D., Holmes, HE., Yu, Y., O’Callaghan, J., Cleary, JO., Sinclair, B., Wiseman, FK., Tybulewicz, VLJ., Fisher, EMC., Lythgoe, MF., Ourselin, S. (2016). Fully-automated µMRI morphometric phenotyping of the Tc1 mouse model of Down syndrome. PLoS ONE 11(9): e0162974. doi:10.1371/journal.pone.0162974
- Holmes, HE., Colgan, N., Ismail, O., Ma, D., Powell, NM., O’Callaghan, JM., Harrison, IF., Johnson, RA., Murray, TK., Ahmed, Z., Heggenes, M., Fisher, A., Cardoso, MJ., Modat, M., Walker-Samuel, S., Fisher, EMC., Ourselin, S., O’Neill, MJ., Wells, JA., Collins, EC., Lythgoe, MF. (2016). Imaging the accumulation and suppression of tau pathology using multi-parametric MRI. Neurobiology of Aging. doi:10.1016/j.neurobiolaging.2015.12.001
- Wells, JA., O’Callaghan, JM., Holmes, HE., Powell, NM., Johnson, RA., Siow, B., Torrealdea, F., Ismail, O., Walker-Samuel, S., Golay, X., Rega, M., Richardson, S., Modat, M., Cardoso, MJ., Ourselin, S., Schwarz, AJ., Ahmed, Z., Murray, TK., O’Neill, MJ., Collins, EC., Colgan, N., Lythgoe, MF. (2015). In vivo imaging of tau pathology using multi-parametric quantitative MRI. NeuroImage, 111, 369–378. doi:10.1016/j.neuroimage.2015.02.023
Links
CMIC software
- NiftyReg, for registration
- NiftySeg, for segmentation
- Mouse brain parcellation
- more Nifty tools
Mouse atlases
Multi-subject atlases are preferred.
© 2014 Nick Powell and University College London, UK. License.