neurotic: NEUROscience Tool for Interactive Characterization
neurotic: NEUROscience Tool for Interactive Characterization
Curate, visualize, annotate, and share your behavioral ephys data using Python
| Distributions: |    |
|---|---|
| Source Code: |   
|
| Tests Status: |   |
| Citing: | 

|
Documentation | Release Notes | Issue Tracker
neurotic is an app for Windows, macOS, and Linux that allows you to easily review and annotate your electrophysiology data and simultaneously captured video. It is an easy way to load your Neo-compatible data (see neo.io for file formats) into ephyviewer without doing any programming. Share a single metadata file with your colleagues and they too will quickly be looking at the same datasets!
To use the app, first organize your datasets in a metadata file like this (see Configuring Metadata):
my favorite dataset:
description: This time it actually worked!
data_dir: C:\local_dir_containing_files
remote_data_dir: http://myserver/remote_dir_containing_downloadable_files # optional
data_file: data.axgx
video_file: video.mp4
# etc
video_offset: -3.4 # seconds between start of video and data acq
epoch_encoder_possible_labels:
- label01
- label02
plots:
- channel: I2
ylim: [-30, 30]
- channel: RN
ylim: [-60, 60]
# etc
filters: # used only if fast loading is off (lazy=False)
- channel: Force
lowpass: 50
# etc
amplitude_discriminators: # used only if fast loading is off (lazy=False)
- name: B3 neuron
channel: BN2
units: uV
amplitude: [50, 100]
# etc
another dataset:
# etcOpen your metadata file in neurotic and choose a dataset. If the data and video files aren't already on your local computer, the app can download them for you, even from a password-protected server or from Google Drive. Finally, click launch and the app will use a standard viewer layout to display your data to you using ephyviewer.
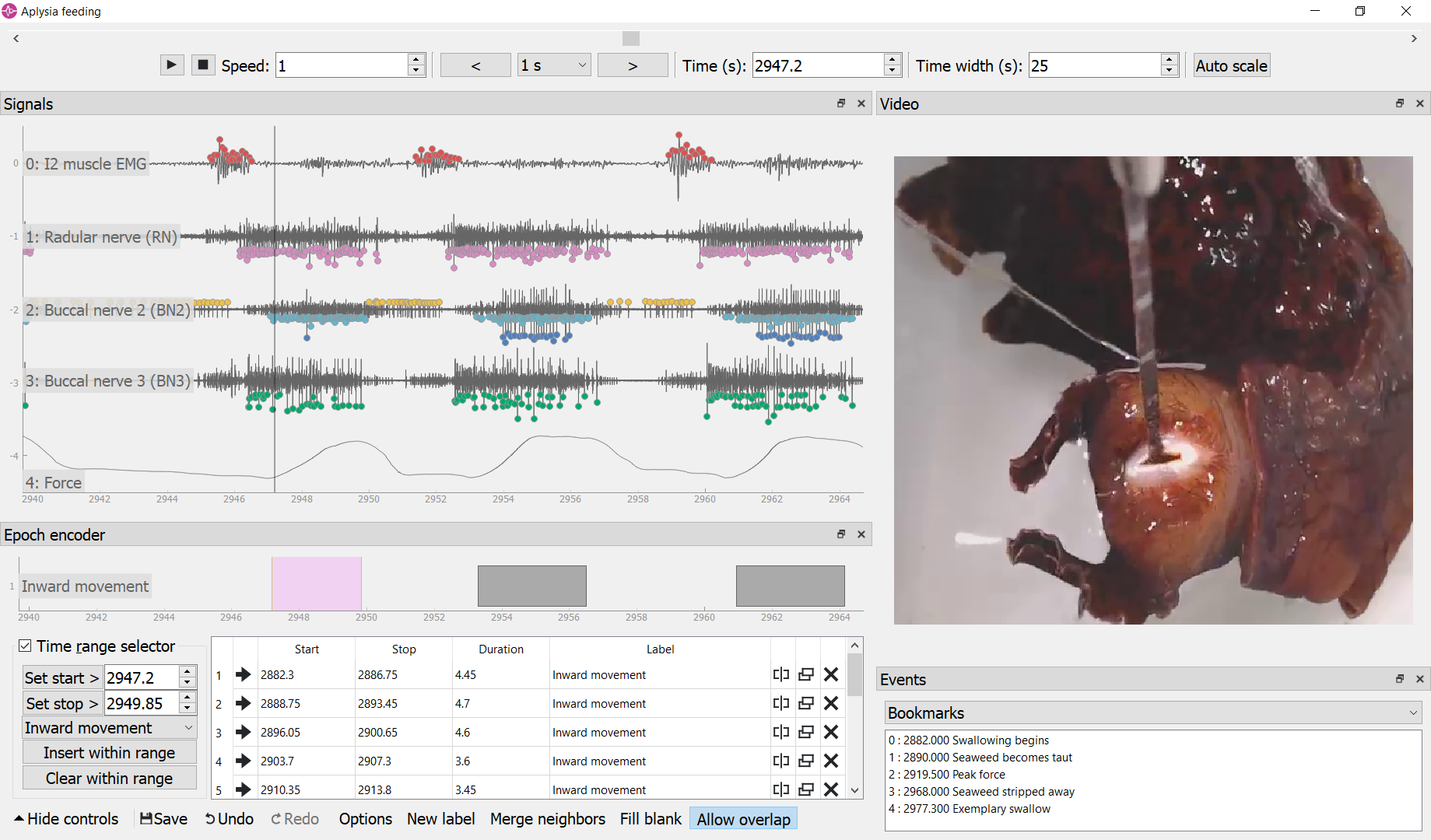
In the screenshot above, the video frame shows a voracious sea slug (Aplysia californica) swallowing a strip of unbreakable seaweed attached to a force transducer. Implanted electrodes recorded from a muscle and the major nerves controlling feeding. The epoch encoder was used to mark the times when seaweed moved into the mouth. Spikes corresponding to activity of identified neurons were detected by neurotic using customizable parameters.
The viewers are easy and intuitive to navigate (see User Interface):
- Pressing the play button will scroll through your data and video in real time, or at a higher or lower rate if the speed parameter is changed.
- The arrow/WASD keys allow you to step through time in variable increments.
- Jump to a time by clicking on an event in the event list or a table entry in the epoch encoder.
- To show more or less time at once, right-click and drag right or left to contract or expand time.
- Scroll the mouse wheel in the trace viewer or video viewer to zoom.
- The epoch encoder can be used to block out periods of time during which something interesting is happening for later review or further analysis (saved to a CSV file).
- All panels can be hidden, undocked, stacked, or repositioned on the fly.
Electrophysiologists will find this tool useful even if they don't need the video synchronization feature!
Portability is easy with neurotic! Use relative paths in your metadata file
along with a remotely accessible data store such as GIN or a Shared Drive on
Google Drive to make your metadata file fully portable. The same metadata file
can be copied to a different computer, and downloaded files will automatically
be saved to the right place. Data stores can be password protected and
neurotic will prompt you for a user name and password. This makes it easy to
share the neurotic experience with your colleagues!
Installing neurotic
neurotic requires Python 3.6 or later.
Standalone Installers (recommended for beginners)
Downloadable installers make installing neurotic easy for beginners. They can be downloaded from the GitHub Releases page:
👉 Download installers here (listed under "Assets")👈
These installers are intended for users who do not want to independently install Python or conda just to use neurotic. They will install neurotic and everything it needs (including a fully contained Python environment) into a dedicated directory on your computer. On Windows, the installer will also create a Start Menu shortcut for launching the app.
For developers, a recipe for building new installers using conda constructor is maintained here: constructor recipe.
Alternate Method: conda (recommended for Pythonistas)
conda users can install neurotic and all of its dependencies with one command:
conda install -c conda-forge neurotic
On Windows, this will also create a Start Menu shortcut for launching the app.
Alternate Method: pip
Install neurotic from PyPI using
pip install neurotic
Note that installation via pip skips one dependency: PyAV, which is
required for displaying videos, and without which neurotic will ignore
videos. PyAV is not easily installed with pip on some systems, especially
Windows. The easiest way to separately install PyAV is using conda:
conda install -c conda-forge av
Updating neurotic
The recommended method of updating neurotic depends on the original method of installation.
If you are unsure what method you used, updating using conda or pip is
likely to work. Standalone installers may be safe too, though this could lead
to having multiple version installed simultaneously.
Updating with Standalone Installers
If you previously installed neurotic using a standalone installer, you may install a newer version using another installer, either into a different directory or by first uninstalling the old version. Installers can be downloaded from the GitHub Releases page:
👉 Download installers here (listed under "Assets")👈
Alternatively, if a new installer is not currently available for your platform, or if you would just like a much faster method, you may use the command line tools provided by the installer (via the "Anaconda Prompt" on Windows, or the Terminal on macOS and Linux):
conda update -c conda-forge neurotic
Updating with conda
If you installed neurotic with conda, you can update to the latest release using
conda update -c conda-forge neurotic
Updating with pip
If you installed neurotic using pip, you can update to the latest release
available on PyPI using
pip install -U neurotic
Development Version
If you are interested in trying new, unreleased features of neurotic, you may install the latest development version from GitHub using
pip install -U git+https://github.com/jpgill86/neurotic.git
Note that if you install the development version, you may also need the latest development version of ephyviewer, which you can get using
pip install -U git+https://github.com/NeuralEnsemble/ephyviewer.git
Getting Started
Windows users who installed using a standalone installer or conda should be able to launch neurotic from the Start Menu.
Mac and Linux users, as well as Windows users, can use the Terminal, command line, or Anaconda Prompt to start the app:
Depending on your operating system, installation method, and environment settings, you may be able to just launch the app from the command line by invoking its name:
neurotic
If the command is not recognized, you likely need to first activate the conda environment into which the app was installed:
conda activate <environment name>
If you used a standalone installer, the environment name may be "
neurotic", so you would useconda activate neurotic
You can then try again invoking the app name:
neurotic
Several examples are provided. Select one, download the associated data using the "Download data" menu action, and then click "Launch". See User Interface for help with navigation.
Disabling "Fast loading" before launch will enable additional features including amplitude-threshold spike detection and signal filtering.
To inspect the metadata file associated with the examples or to make changes to it, click "Edit metadata". See Configuring Metadata for details about the format.
If you like working with Jupyter notebooks, you can launch an example notebook that includes a tutorial for using neurotic's API:
neurotic --launch-example-notebook
The command line interface accepts other arguments too:
usage: neurotic [-h] [-V] [--debug | --no-debug] [--lazy | --no-lazy]
[--thick-traces | --no-thick-traces]
[--show-datetime | --no-show-datetime]
[--ui-scale {tiny,small,medium,large,huge}]
[--theme {light,dark,original,printer-friendly}]
[--use-factory-defaults] [--launch-example-notebook]
[file] [dataset]
neurotic lets you curate, visualize, annotate, and share your behavioral ephys
data.
positional arguments:
file the path to a metadata YAML file (default: an example
file)
dataset the name of a dataset in the metadata file to select
initially (default: the first entry in the metadata
file)
optional arguments:
-h, --help show this help message and exit
-V, --version show program's version number and exit
--debug enable detailed log messages for debugging
--no-debug disable detailed log messages for debugging (default)
--lazy enable fast loading (default)
--no-lazy disable fast loading
--thick-traces enable support for traces with thick lines, which has
a performance cost
--no-thick-traces disable support for traces with thick lines (default)
--show-datetime display the real-world date and time, which may be
inaccurate depending on file type and acquisition
software
--no-show-datetime do not display the real-world date and time (default)
--ui-scale {tiny,small,medium,large,huge}
the scale of user interface elements, such as text
(default: medium)
--theme {light,dark,original,printer-friendly}
a color theme for the GUI (default: light)
--use-factory-defaults
start with "factory default" settings, ignoring other
args and your global config file
alternative modes:
--launch-example-notebook
launch Jupyter with an example notebook instead of
starting the standalone app (other args will be
ignored)
Defaults for arguments and options can be changed in a global config file,
.neurotic\neurotic-config.txt, located in your home directory.
Citing neurotic
To cite neurotic in your publication, please refer to:
Gill, J. P., Garcia, S., Ting, L. H., Wu, M., & Chiel, H. J. (2020). neurotic: Neuroscience Tool for Interactive Characterization. eNeuro, 7(3). https://doi.org/10.1523/ENEURO.0085-20.2020
Specific versions of the software can be cited from archives at Zenodo.
Documentation
For detailed information on configuring metadata, working examples, the API reference guide, release notes, and more, see the Documentation.